Columns used to define observations
OBSERVATION: response
The OBSERVATION column-type can be used to record continuous (PKanalix and Monolix), categorical (Monolix), count (Monolix) or time-to-event (Monolix) data. For dose lines, the content is free and will not be used. For response lines, the requirements depend on the type of data and are summarized below.
Continuous data
The value represents what has been measured (e.g concentrations) and can be any double value.
Examples
Basic example:
ID TIME AMT Y
1 0 50 .
1 0.5 . 1.1
1 1 . 9.2
1 1.5 . 8.5
1 2 . 6.3
1 2.5 . 5.5Full data set for continuous data: theophylline data set, the warfarin data set, and the HIV data set among others can be found here.
Categorical data
https://www.youtube.com/watch?v=ZG7LhmApb_sIn case of categorical data, the observations at each time point can only take values in a fixed and finite set of nominal categories. In the data set, the output categories must be coded as consecutive integers.
Examples
Basic example:
ID TIME Y
1 0.5 3
1 1 0
1 1.5 2
1 2 2
1 2.5 3Full data set for joint continuous and categorical data: One can see the respiratory status data set and the warfarin data set for example for more practical examples on a categorical and a joint continuous and categorical data set respectively.
Count data
Count data can take only non-negative integer values that come from counting something, e.g., the number of trials required for completing a given task. The task can for instance be repeated several times and the individuals performance followed.
Count data can also represent the number of events happening in regularly spaced intervals, e.g the number of seizures every week. If the time intervals are not regular, the data may be considered as repeated time-to-event interval censored, or the interval length can be given as regressor to be used to define the probability distribution in the model.
Examples
Basic example: in the data set below, 10 trials are necessary the first day (t=0), 6 the second day (t=24), etc.
ID TIME Y
1 0 10
1 24 6
1 48 5
1 72 2One can see the epilepsy attacks data set for a more practical example.
(Repeated) time-to-event data
https://youtu.be/E3oHXlbQU0gIn this case, the observations are the “times at which events occur“. An event may be one-off (e.g., death) or repeated (e.g., epileptic seizures, mechanical incidents, strikes). In addition, an event can be exactly observed, interval censored or right censored.
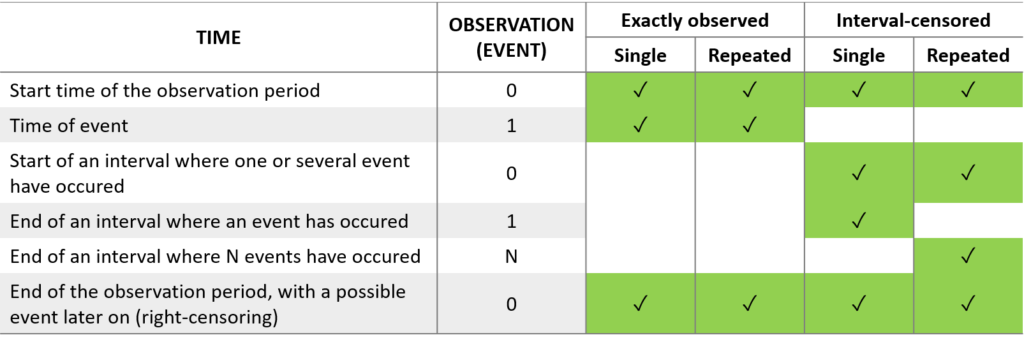
For the formatting of time-to-event data, the column TIME should contain:
the time of an event,
the time at which the observation period starts (required by the MonolixSuite, contrary to other tools for survival analysis. This allows to define the data set using absolute times, in addition to durations (if the start time is zero, the records represent durations between the start time and the event).
the end of the observation period or time intervals for interval-censoring.
The column OBSERVATION contains an integer that indicates how to interpret the associated time. The different values for each type of event and observation are summarized in the table below:
The figure below summarizes the different situations with examples:
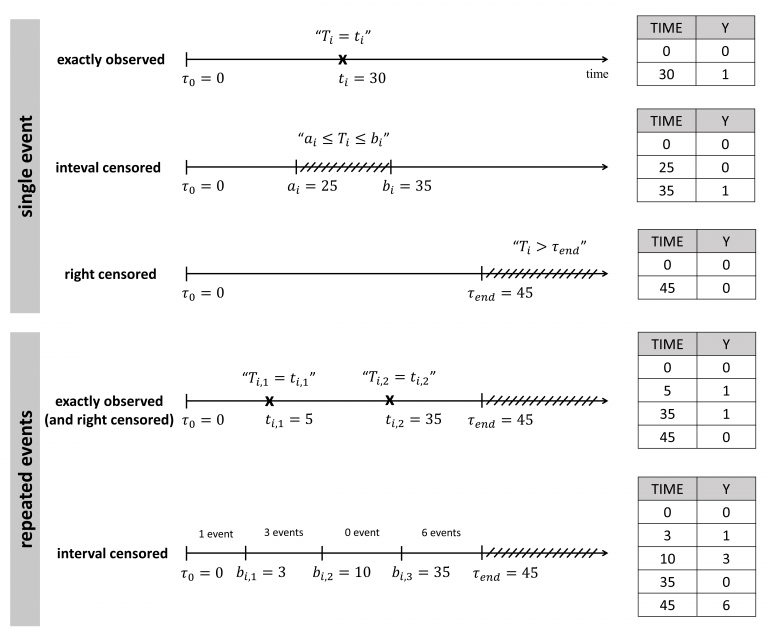
Single events exactly observed data
One must indicated the start time of the observation period with Y=0, and the time of event (Y=1) or the time of the end of the observation period if no event has occurred (Y=0).
Examples
Basic example: in the following dataset, the observation period last from starting time t=0 to the final time t=80. For individual 1, the event is observed at t=34, and for individual 2, no event is observed during the period. Thus it is noticed that at the final time (t=80), no event occurred.
ID TIME Y
1 0 0
1 34 1
2 0 0
2 80 0Using absolute times instead of duration, we could equivalently write:
ID TIME Y
1 20 0
1 54 1
2 33 0
2 113 0The duration between start time and event (or end of the observation period) are the same as before, but this time we record the day at which the patients enter the study and the days at which they have events or leave the study. Different patients may enter the study at different times.
Full data sets for time-to-event data: PBC data set and Oropharynx data set
Repeated events exactly observed data
One must indicate the start time of the observation period (Y=0), the end time (Y=0) and the time of each event (Y=1).
Examples
Basic example: below the observation period last from starting time t=0 to the final time t=80. For individual 1, two events are observed at t=34 and t=76, and for individual 2, no event is observed during the period.
ID TIME Y
1 0 0
1 34 1
1 76 1
1 80 0
2 0 0
2 80 0Single events interval censored data
When the exact time of the event is not known, but only an interval can be given, the start time of this interval is given with Y=0, and the end time with Y=1. As before, the start time of the observation period must be given with Y=0.
Examples
Basic example: we only know that the event has happened between t=32 and t=35.
ID TIME Y
1 0 0
1 32 0
1 35 1Repeated events interval censored data
In this case, we do not know the exact event times, but only the number of events that occurred for each individual in each interval of time. The column-type Y can now take integer values greater than 1, if several events occurred during an interval.
Examples
Basic example: No event occurred between t=0 and t=32, 1 event occurred between t=32 and t=35, 1 between t=35 and t=50, none between t=50 and t=56, 2 between t=56 and t=78 and finally 1 between t=78 and t=80.
ID TIME Y
1 0 0
1 32 0
1 35 1
1 50 1
1 56 0
1 78 2
1 80 1Warnings
If a subject or a subject-occasion has no observations, a warning message arises telling which subjects or subjects-occasions have no measurements and will be ignored.
Format restrictions
A data set shall not contain more than one column with column-type OBSERVATION. Multiple observation types need to be distinguished with the OBSERVATION ID column.
If EVENT ID = 1, 3 or 4, the content of the OBSERVATION column is free and ignored.
If EVENT ID = 0, and IGNORED OBSERVATION = 1 or 2, the content of the OBSERVATION column is free and ignored.
If EVENT ID = 0 and IGNORED OBSERVATION = 0, the contant of the OBSERVATION column has to be a double (dots ‘.’ are not allowed).
If the EVENT ID column is absent (and whatever the IGNORED OBSERVATION column is), the content is free. Values that are not doubles (for instance dots ‘.’ or strings) are ignored.
OBSERVATION ID: response identifier
The OBSERVATION ID column permits to distinguish several types of observations (several concentrations, effects, etc). The OBSERVATION ID column assigns an identifier to each observation of the OBSERVATION column. Those identifiers are used to map the observations of the data set to the outputs of the model (in the OUTPUT block of the model file). The dot “.” is not considered as a repetition of the previous line but as a different identifier.
There can be more OBSERVATION ID values than there are outputs in the model file. In that case only the observations corresponding to the first identifiers(s) in alphabetical order will be used (example below).
For continuous data, in the Monolix graphical user interface, the data viewer and the plots, the observations will be called yX with X corresponding to the identifier given in the OBSERVATION ID column (for instance y1 and y2 if identifiers 1 and 2 were used in the OBSERVATION ID column).
https://youtu.be/Z2e2YOJYNtQExamples
Basic example with integers: with the following data set
TIME AMT OBS YTYPE
0 . 12 1
5 . 6 2
10 . 4 1
15 . 3 2and the following output block
OUTPUT:
output = {Cc, R}the observations “12” and “4” which have identifier “1” will be matched to the output “Cc”, while observations ”6″ and “3” with identifier “2” will be matched to “R”.
Basic example with strings (not recommended): with the following data set
TIME AMT OBS YTYPE
0 . 12 PK
5 . 6 PK
10 . 4 PD
15 . 3 PDand the following OUTPUT block in the Mlxtran model file:
OUTPUT:
output = {Cc, R}the observations tagged with “PD” will be mapped to the first output “Cc” (which is probably not what is desired), and those tagged with “PK” will be mapped to the second output “R”, because in alphabetical order “PD” comes before “PK”.
Basic example with more OBSERVATION ID values than model outputs: with the following data set
TIME AMT OBS YTYPE
0 . 12 1
5 . 6 2
10 . 4 1
15 . 3 2and the following output block
OUTPUT:
output = {Cc}the observations tagged with identifier “1” will be mapped to the model output “Cc” and the observations tagged with “2” will be ignored. If the user wants to use only the data tagged with “2”, he can add an IGNORED OBSERVATION column which ignores all observations with identifier “1” (see below).
Basic example with more OBSERVATION ID values than model outputs and an IGNORED OBSERVATION column: with the following data set
TIME AMT OBS YTYPE MDV
0 . 12 1 1
5 . 6 2 0
10 . 4 1 1
15 . 3 2 0and the following output block
OUTPUT:
output = {Cc}the observations tagged with identifier “1” will all be ignored (due to the MDV column tagged as IGNORED OBSERVATION) and the observations with identifier “2” will be mapped to the model output “Cc”.
Full data set example: warfarin PKPD data set, PSA and survival data set
Format restrictions
A data set shall not contain more than one column with column-type OBSERVATION ID.
The content of the OBSERVATION ID column can be strings or integers.
The dot “.” is not considered as a repetition of the previous line but as a different identifier.
CENSORING: censored observation
The CENSORING column permits to mark censored data. When an observation is marked as censored, the (upper or lower) limit of quantification is given in the OBSERVATION column (not in a separate column).
CENSORING = 1 means that the value in OBSERVATION column is a lower limit of quantification (LLOQ). The true observation y verifies y<LLOQ.
CENSORING = 0 means the value in response-column corresponds to a valid observation (no interval associated).
CENSORING = -1 means that the value in OBSERVATION column is an upper limit of quantification (ULOQ). The true observation y verifies y>ULOQ.
The mathematical handling of censored data is described here.
https://youtu.be/pEQYCva3CWcFormat restrictions
A data set shall not contain more than one column with column-type CENSORING.
For dose lines, the content is free and will be ignored.
For response lines, there are only four possible values : -1, 0, 1 and ‘.’ (interpreted as 0).
LIMIT: limit for censored values
When the column LIMIT contains a numeric value and CENSORING is different from 0, the value in the LIMIT column is interpreted as the second bound of the interval. Writing yobs the value in the OBSERVATION column and ylimit the value in the LIMIT column, the true observation y verifies y∈[ylimit,yobs]. When LIMIT = ‘.’ , the value is interpreted as -infinity or +infinity depending on the value of CENSORING (1 or -1 respectively) as if the LIMIT column would not be present.
Format restrictions
A data set shall not contain more than one column with column-type LIMIT.
A data set shall not contain any column with column-type LIMIT if no column with column-type CENSORING is present.
Allowed values are doubles and dot ‘.’ , and strings are not allowed (even when CENSORING = 0).
Example with CENSORING and LIMIT
It is possible to have both censoring type on the same individual, i.e. both upper or lower limit of quantification. It is possible to have measurements with and without bounds. The example below gives an overview of the possible combinations:

