Introduction: model structure for Monolix
Structural model
In Monolix, the model code defined in the “Structural model” tab (external .txt file loaded into Monolix or model selected from a library) contains only the structural part of the model in a single [LONGITUDINAL] section.
The [LONGITUDINAL] section is itself divided into blocks, which allows to define different types of variables in different ways. Here are the possible blocks and variable lists in that section:
DESCRIPTION: (optional): block to put comments on the model
input (mandatory): list of input parameters
PK: (optional): block were macros can be used and combined to define compartments and transfers between them
EQUATION: (optional): block were ODEs and DDES can be defined, as well as mathematical expressions (e.g parameter transformations)
DEFINITION: (optional): block to define random variables (discrete or continuous) which represent observations, via their probability distribution: error model, probability distribution for count/categorical output and hazard for time-to-event output
OUTPUT: (mandatory): block indicating the output of the model
output (mandatory): list of output parameters included in the OUTPUT block.
In all blocks, it is possible to use:
Reserved keywords of the Mlxtran language cannot be used as names for other parameters. Mlxtran is a declarative language, not an imperative language. Therefore equations are mathematical definitions rather than a series of instructions.
Inputs
In Monolix, the input = { } list of the [LONGITUDINAL] section declares the individual parameters and the regressors.
[LONGITUDINAL]
input = {ka, V, Cl}For regressors, the regressor status must be specified using the following syntax, for instance for two regressors called regvar1 and regvar2. One line per regressor is necessary. As a reminder, in Monolix, the regressors are mapped to the regressor columns of the data set by declaration order (first column tagged as regressor mapped to the first regressor appearing in the input list).
[LONGITUDINAL]
input = {..., regvar1, regvar2}
regvar1 = {use=regressor}
regvar2 = {use=regressor}Example:
In this example, ka, V, Cl, Emax and EC50 are individual parameters and E0 is declared as a regressor and will be read from the first data set column tagged as regressor.
[LONGITUDINAL]
input = {ka, V, Cl, E0, Emax, EC50}
E0 = {use = regressor}Outputs
In Monolix, the OUTPUT: block declare the variables which will be mapped to the observations in the data set via the output={} list, and the additional variables recorded in the output tables via the table={} list. The outputs are mapped to the observation ids of the data set via the mapping panel in the Monolix GUI. The variables in the table={} statement are outputted in the result folder of Monolix.
Example:
OUTPUT:
output = {Conc, Effect}
table = {Ap, T12}Library of models and examples
The MonolixSuite contains libraries of pre-written structural models which can be directly selected via the GUI in Monolix:
PK: typical pharmacokinetics models with several types of administrations and 1 to 3 compartments
PK/PD: joint PK/PD model with direct effect, effect compartment or turnover
TMDD: target mediated drug disposition models
Parent-Metabolite models
TTE: time-to-event models
Count: models for count data
TGI: tumor growth and tumor growth inhibition models
Examples of common models which are not (yet) in the libraries are also given here: Advanced models
Statistical model
In Monolix the statistical part of the model (covariate effects, parameter distributions and error model) is defined through the graphical user interface. Then when saving a Monolix project, the statistical part of the model is automatically written in the .mlxtran project file using mlxtran language, divided into three parts:
The observation (error) model: defined inside the DEFINITION: block below the [LONGITUDINAL] section.
The individual model: describes the distributions for the individual parameters, correlation structure and covariate effects, defined inside the [INDIVIDUAL] section.
The covariates defined in the project: described inside the [COVARIATE] section.
Example
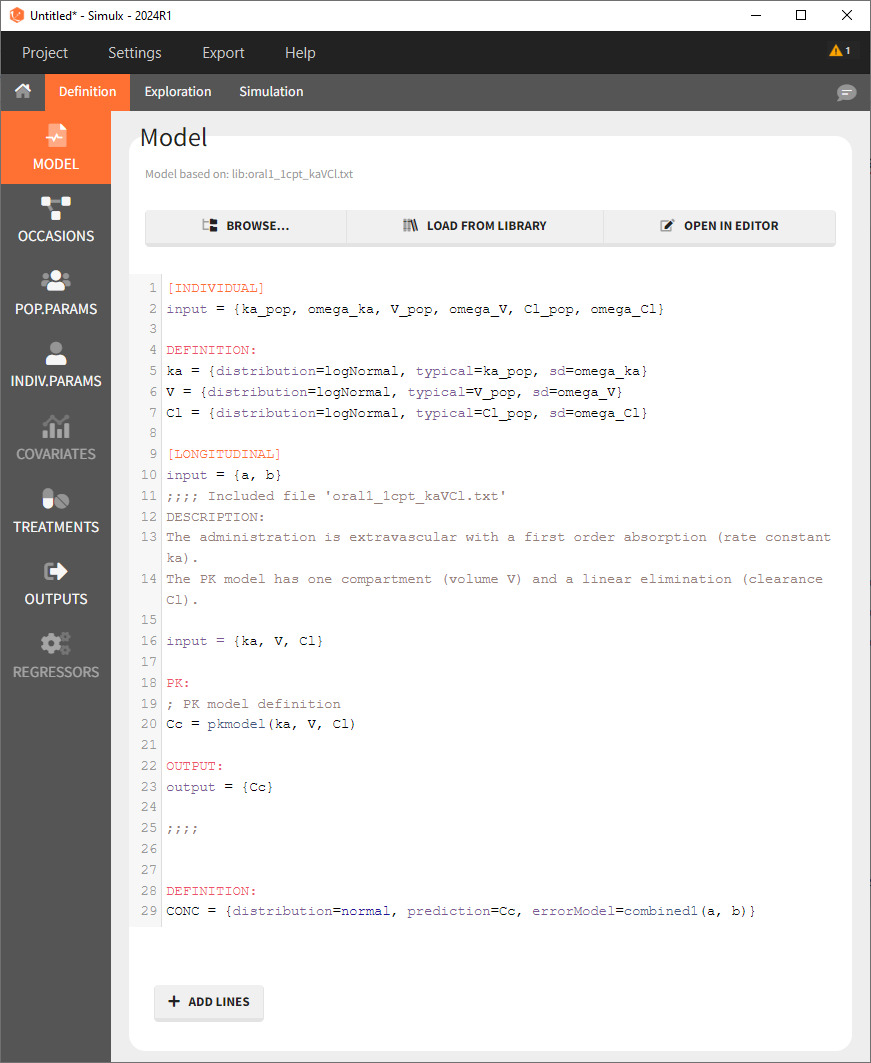
Here is the description of the full model (structural and statistical) corresponding to the Monolix demo 1.1.libraries_of_models/theophylline_project.mlxtran, automatically saved by Monolix inside the <MODEL> part of the theophylline_project.mlxtran file:
<MODEL>
[COVARIATE]
input = {WEIGHT, SEX}
SEX = {type=categorical, categories={'F', 'M'}}
[INDIVIDUAL]
input = {ka_pop, omega_ka, V_pop, omega_V, Cl_pop, omega_Cl}
DEFINITION:
ka = {distribution=logNormal, typical=ka_pop, sd=omega_ka}
V = {distribution=logNormal, typical=V_pop, sd=omega_V}
Cl = {distribution=logNormal, typical=Cl_pop, sd=omega_Cl}
[LONGITUDINAL]
input = {a, b}
file = 'lib:oral1_1cpt_kaVCl.txt'
DEFINITION:
CONC = {distribution=normal, prediction=Cc, errorModel=combined1(a, b)}Note that when exporting the model from Monolix to Simulx, this full model then appears in the interface of Simulx: