Lifespan based indirect response (LIDR) models have originally been proposed in the field of hematology. The population of cells are described by cell production and cell degradation. The main assumption of LIDR models is that a cell dies when its lifespan expires. This implies that the cell degradation rate is equal to the cell production rate delayed by the lifespan. This assumption differs from classical indirect response model where the cell (or response) degradation is proportional to the cell number (first-order process), independently of the production rate or the lifespan.
LIDR models have been reviewed in detail in:
The implementation of LIDR model makes use of delayed differential equations, which can easily be solved in the MonolixSuite using the delay differential equation solver. In the following, we will consider a drug with simple exponential decay after a bolus administration and a response R, which can for instance be the number of blood cells.
Mlxtran code for LIDR models
LIDR model for drugs affecting the production rate of the response
In this model, the drug affects the response via an Imax model on the production rate. The equation for the response is:
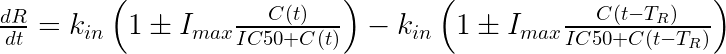
where + would be used if the drug increases the production rate and – if it decreases it. Notice that the degradation rate is simply the production rate delayed by the lifespan
. If necessary, a sigmoidicity parameter gamma can be added to the formula. When implemented in mlxtran,
translates into delay(C,TR) which means that we use the value of the concentration C delayed by time
.
Note that the delay function makes use of the DDE solver, which is slower than the ODE solver. This video presents an alternative implementation using ODEs only, which leads to greatly shorter run times.
The mlxtran code reads (for the inhibition case):
[LONGITUDINAL]
input={V, k, kin, Imax, IC50, TR}
PK:
depot(target=Ac)
EQUATION:
t_0 = 0
Ac_0 = 0
R_0 = kin * TR
Cc = Ac/V
ddt_Ac = -k*Ac
ddt_R = kin*(1 - Imax *Cc/(Cc+IC50)) - kin*(1 - Imax *delay(Ac,TR)/(delay(Ac,TR)+IC50*V))
OUTPUT:
output = {Cc, R}
As usual, the depot macro permits to link the doses defined in the data set to the DDEs system.
In the model formulation above, it is assumed that both the drug concentration Cc and the response R are recorded in the data set. If only R is observed, the output line should be changed to output= {R}. Note that if the concentration Cc is not observed, it is not possible to identify both V and IC50. The value of V can then be fixed. Alternatively, the K-PD model can be reformulated as in Jacqmin et al. (2007).
In the delay macro, it is mandatory to use an ODE variable. We thus use Ac rather than Cc and divide by the volume V outside of the delay macro.
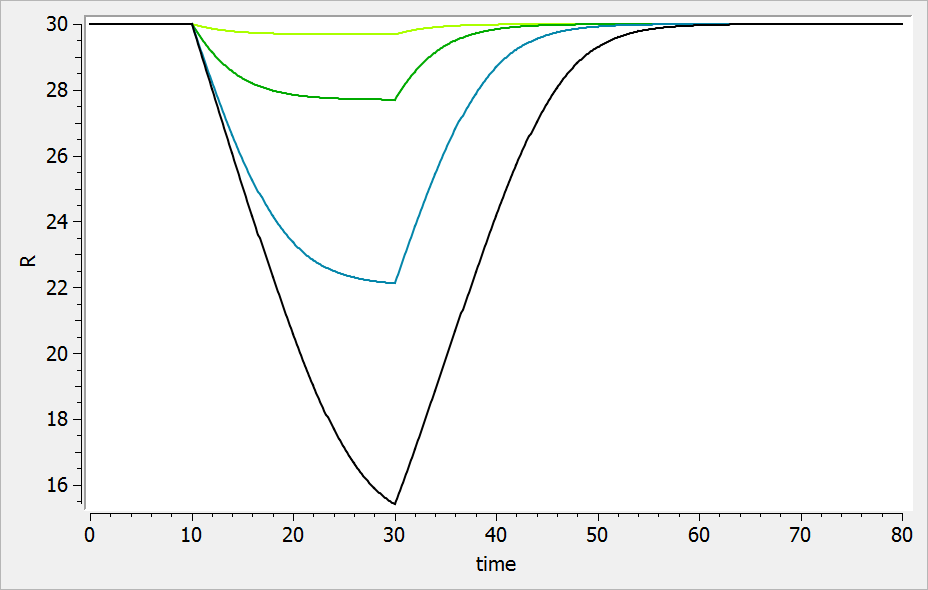
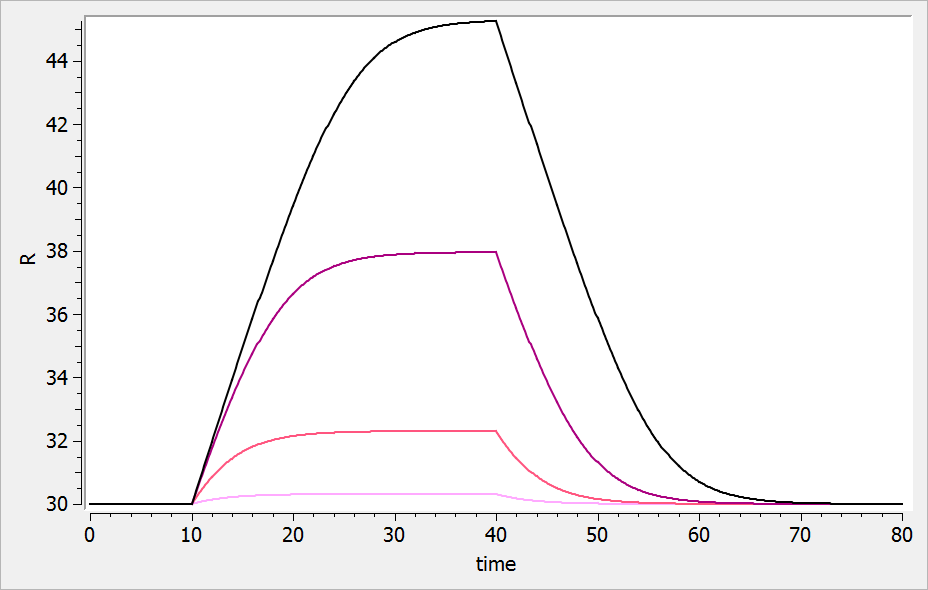
For a drug inhibiting the production rate, the typical response shape is the following (darker colors
indicate larger drug amounts). The response start to change from baseline at time 0 and is maximal at time
:
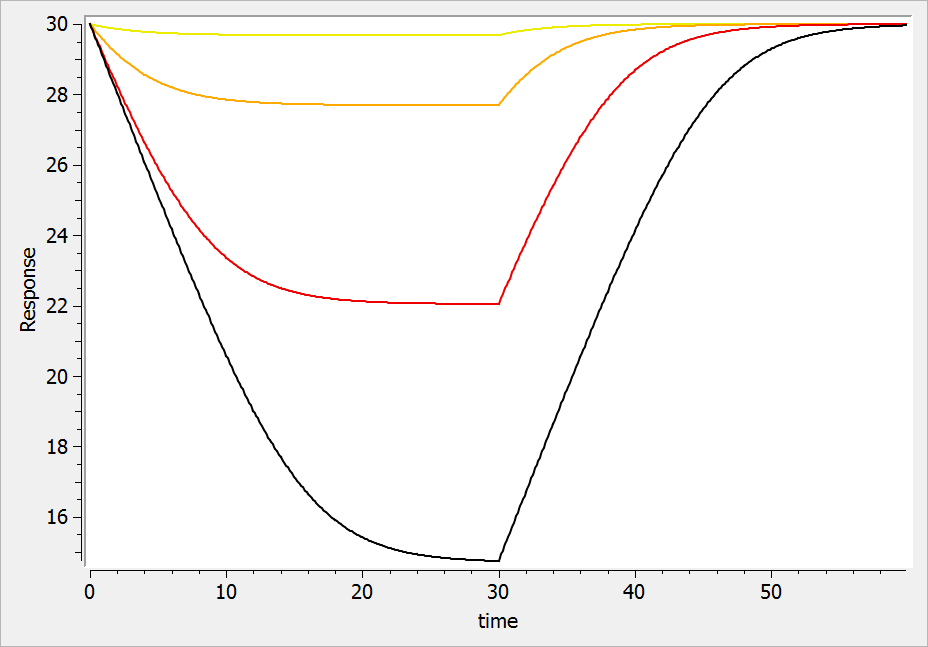
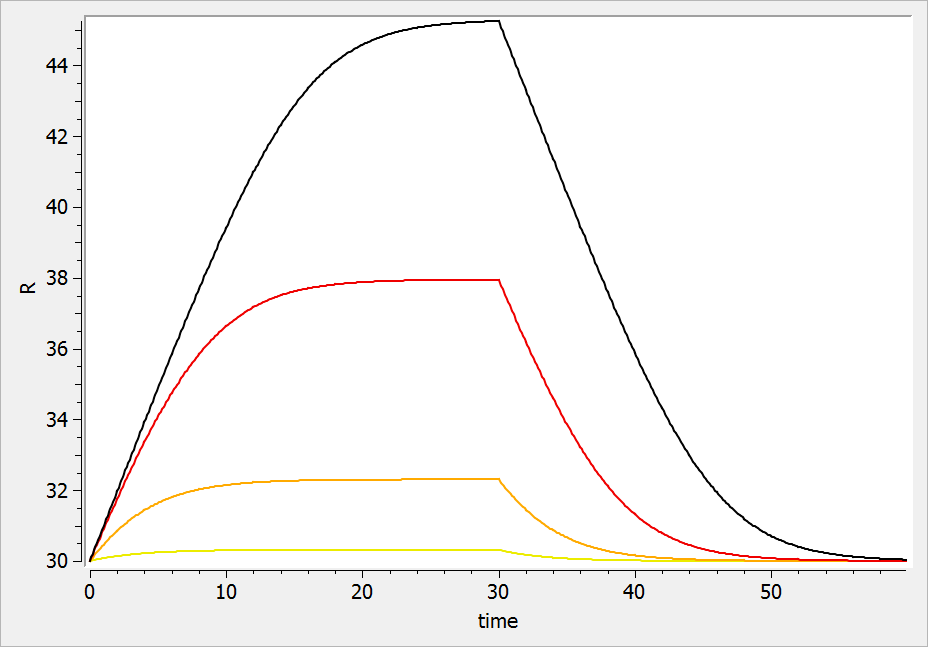
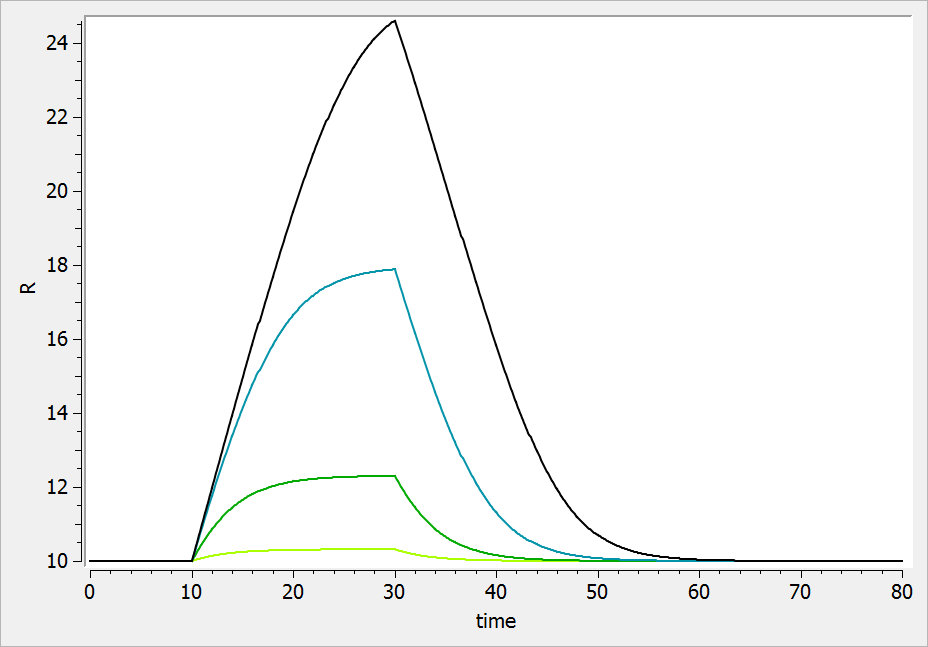
If the drug would increase the production rate instead of decreasing it, the response would increase over time before recovering its basal level:
LIDR model for drugs affecting the cell lifespan distribution
This model assumes that initially the lifespan is
and that the drugs modifies the cell lifespan to a new value
. The drug effect is described by an Imax term, which represents the fraction of cells with lifespan
. The higher the drug concentration is, the more cells will switch from the baseline lifespan
to the new lifetime
.
The equation for the response is:
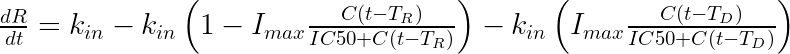
The mlxtran code reads:
[LONGITUDINAL]
input={V, k, kin, Imax, IC50, TR, TD}
PK:
depot(target=Ac)
EQUATION:
t_0 = 0
Ac_0 = 0
R_0 = kin * TR
Cc = Ac/V
ddt_Ac = -k*Ac
ddt_R = kin - kin*(1 - Imax *delay(Ac,TR)/(delay(Ac,TR)+IC50*V)) - kin*(Imax *delay(Ac,TD)/(delay(Ac,TD)+IC50*V))
OUTPUT:
output = {Cc, R}
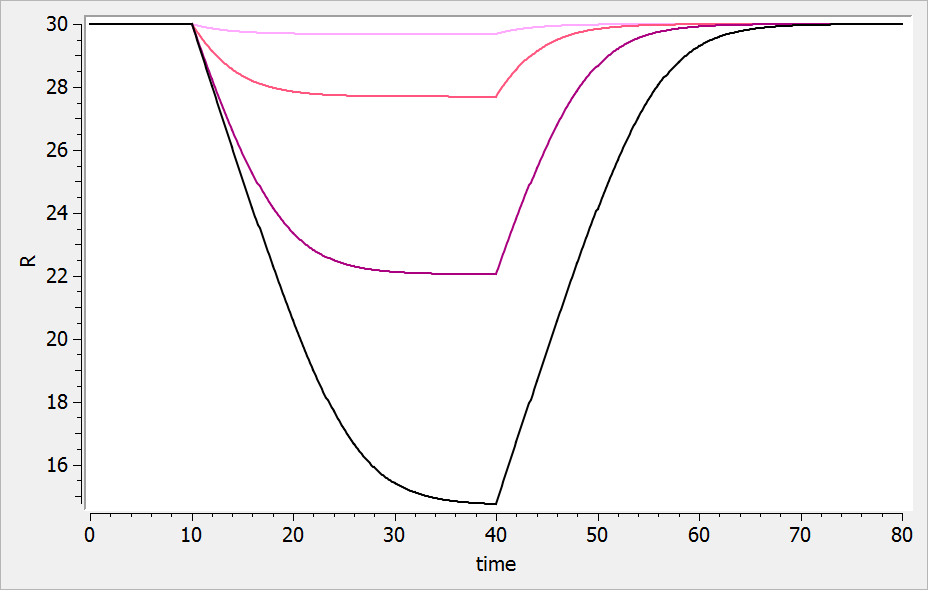
For a drug reducing the lifespan from
to
days, the typical response shape is the following (darker colors indicate larger drug amounts). The response is maximal at time
. Note that there is a delay before the response starts to change, which corresponds to
.
The possibility to have this delay requires one more parameter compared to the lifespan model for drugs affecting the production rate.
If the drug would increase the lifespan (i.e.
), the curves would be flipped over (increasing response):
LIDR model with a precursor pool
In this model we assume that the drug effects a precursor pool, which matures into the main pool represented by the response R. Interestingly, it is not necessary to solve the equation for the precursor, only the equation for the response R is needed:
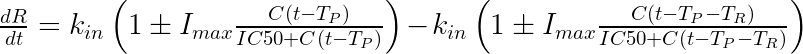
where + is used if the drug increases the precursor production rate and – if it decreases it.
represents the lifespan of the main cells (response) and
the lifespan of the precursor.
The mlxtran code reads:
[LONGITUDINAL]
input={V, k, kin, Imax, IC50, TR, TP}
PK:
depot(target=Ac)
EQUATION:
t_0 = 0
Ac_0 = 0
R_0 = kin * TR
Cc = Ac/V
ddt_Ac = -k*Ac
ddt_R = kin*(1 - Imax *delay(Ac,TP)/(delay(Ac,TP)+IC50*V)) - kin*(1 - Imax *delay(Ac,TP+TR)/(delay(Ac,TP+TR)+IC50*V))
OUTPUT:
output= {Cc, R}
The typical response-time curve is displayed below for the case where the drug inhibits the precursor production. The response start to change at time
and the change is maximal at time
.
If the drug increases the precursor production, we obtain:
Exploration of indirect model and LIDR model differences with Mlxplore
We explore the differences between:
-
the LIDR model
-
the indirect response model
In both models, we assume that the drug inhibits the response production rate. Yet, in the LIDR model, the response degradation rate is equal to the production rate delayed by the lifespan. On the opposite, in the indirect response model, the response degradation rate is proportional to the response magnitude.

