Calculating the half-life
The half-life is usually not part of the estimated parameters but it often needs to be reported. It presents the advantage of having units of time and being easy to understand. The terminal plasma half-life is the time required to divide the plasma concentration by two after reading pseudo-equilibrium. It is especially relevant to multiple dosing regimens, because it controls the degree of drug accumulation and the time taken to reach equilibrium. For a more detailed overview, please refer to:
This documentation page explains the half-life formulas and how to modify the Monolix or Simulx models to calculate it.
Half-life formulas
One-compartmental model
For a drug captured by a model with one compartment and linear elimination, and without flip-flop kinetics, the half-life is calculated as:
|
|
with the elimination rate,
the clearance and
the volume of distribution. Note that
so you might also find the formula written with this value.
Two-compartmental model
For a drug captured by a model with one compartment and linear elimination, the half-life correspond to the slope of the second part of the curve on a log-concentration over time plot, once pseudo-equilibrium is achieved. The slope of the terminal (second) phase is usually called , the first slope
and the intercepts A and B.
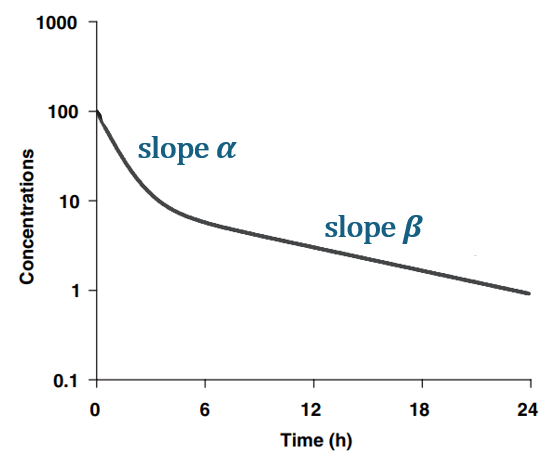
The parametrization of the 2-compartment model with (A, B,) is an alternative parametrization compared to the parametrization with (k, V, k12, k21) or (Cl, V, Q, V2).
It is possible to convert between these different parametrizations using the following formulas:
|
|
The half-life corresponds to the terminal phase and is defined as:
|
|
Implementation in Monolix
The calculation can be added to the structural model and outputted using the table={} statement.
Let’s take the demo project theophylline as an example, which use a model from the PK library with parametrization (ka, V, Cl).
After loading the project, in the Structural model tab, click “EDIT MODEL” and add the following lines and save the file under a new name.
[LONGITUDINAL]
input = {ka, V, Cl}
PK:
; PK model definition
Cc = pkmodel(ka, V, Cl)
Thalf = log(2)*V/Cl
OUTPUT:
output = {Cc}
table = {Thalf}After running the project, the half-life values are displayed in the file tables.txt in the result folder.
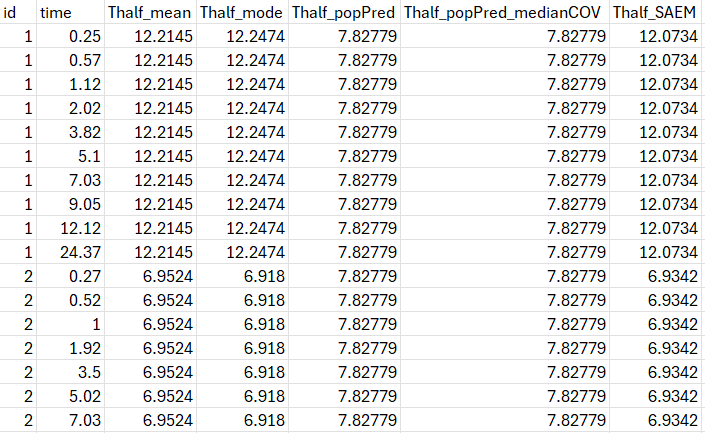
The values will be identical on all lines corresponding to the same individuals (or occasion in case of IOV).
Thalf_mode corresponds to the individual half-life value calculated using the EBEs.
Thalf_popPred corresponds to the value calculated using the population parameters (and individual covariates values if covariates are used on V and Cl)
For a two-compartmental model from the PK library, the lines to add would be:
[LONGITUDINAL]
input = {Cl, V1, Q, V2}
PK:
; Parameter transformations
V = V1
k12 = Q/V1
k21 = Q/V2
; PK model definition
Cc = pkmodel(V, Cl, k12, k21)
k = Cl/V1
beta = 0.5*(k12+k21+k-sqrt((k12+k21+k)^2-4*k21*k))
Thalf = log(2)/beta
OUTPUT:
output = {Cc}
table = {Thalf}