Bootstrap is not available in versions prior to 2024R1
Introduction
The Bootstrap module in Monolix provides a robust method to assess parameter uncertainty, offering an alternative to calculating standard errors via inversion of the Fisher Information Matrix. This approach becomes particularly valuable when facing issues such as NaNs in standard errors due to numerical errors in matrix inversion or biases in results caused by incorrect assumptions of asymptotic normality for parameter estimates. Bootstrap overcomes these challenges by sampling many replicate datasets and re-estimating parameters on each replicate.
While powerful, bootstrap comes with certain drawbacks:
-
Running many replicates for population parameter estimation can be time-consuming. Bootstrap in Monolix can be used from command line and with distributed calculation (parallelization on a grid via MPI).
-
Saving a large number of new datasets and results may raise storage issues. You can choose in the settings whether to save sampled datasets and results.
Accessing the bootstrap module
Bootstrap can be accessed under the “Perspective” menu or with a shortcut next to “Run” in the “Statistical model & Tasks” tab:
Available bootstrap settings
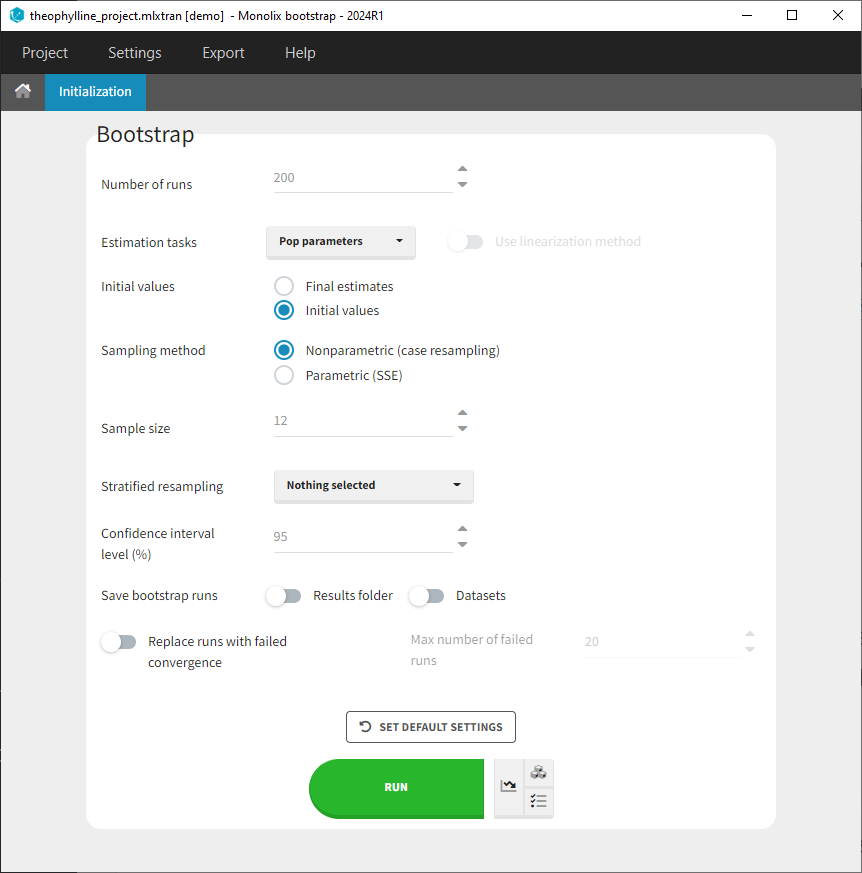
Users can customize the following settings:
-
Number of runs: Number of bootstrap replicates
-
Estimation tasks: Estimation tasks to run among population pramaters, Standard Errors, and Log-Likelihood
-
Initial values: Whether bootstrap runs should start their estimation from the same initial values as the initial run, or from the final estimated values from the initial run.
-
Sampling method: Type of bootstrap:
-
Nonparametric Bootstrap (case resampling): New datasets are sampled from the initial dataset for each replicate. Each individual of the new dataset is sampled randomly with replacement from the initial dataset. This means that some individuals from the original dataset will appear several times in the resampled dataset, while some individuals will not appear at all. The generated datasets are thus similar but different compared to the original dataset. This method makes no assumption on the model.
-
Parametric Bootstrap (SSE): This method is also called SSE for stochastic simulation and estimation. New datasets are simulated from the model for each replicate. Individual parameters are sampled from the population distribution and individuals are simulated using the same design as in the original dataset (same treatments, covariates, regressors, and observation times). Residual error is added on top of the model predictions to obtain a realistic dataset. If the initial dataset has censored data, censoring limits to apply to the simulated datasets can be specified by the user. This method assumes that the model correctly captures the original data.
-
-
Sample size: Number of individuals in the bootstrap datasets. By default it is set equal to the number of individuals of the original dataset but can be modified if required.
-
Stratified resampling: Selection of categorical covariates which distribution should be preserved when re-sampling individuals to create datasets for non-parametric bootstrap. When the original dataset contains few individuals, or when a categorical covariate distribution is highly imbalanced with only few individuals belonging to one of the categories, resampled bootstrap datasets can results in quite different distributions (e.g only one individual left in one of the categories) and this can lead to a bias, in particular on the covariate effects (beta) parameters. To avoid this situation, the sampling can be done such that the number of individuals in each category of the covariates selected in “stratified resampling” remains the same as in the initial dataset.
-
Confidence interval level: Level of the confidence interval bounds displayed in the results to summarize the uncertainty on the population parameters.
-
Save bootstrap runs: Option to save or not save bootstrap datasets and results folder of each bootstrap run. Both options need to be selected if you want to be able to reload each bootstrap run including the results.
-
Replace runs with failed convergence: Option to replace or not bootstrap replicates that have a failed convergence. This option allows to replace runs for which the “auto-stop” criteria of the exploratiry phase of the population parameter estimation have not been reached. You can also set a maximal number of runs to replace, so that bootstrap will not go on forever if many runs do not converge. Bootstrap will stop once the maximal number is reached.
Running bootstrap
After clicking on “Run”, bootstrap is launched and the population parameters estimates from all runs already done are shown as a table and as a plot of their medians and their confidence intervals with respect to the bootstrap iterations. The table and plot updates as soon as new bootstrap run finishes. An estimation of the remaining time to complete all bootstrap runs is available at the top.
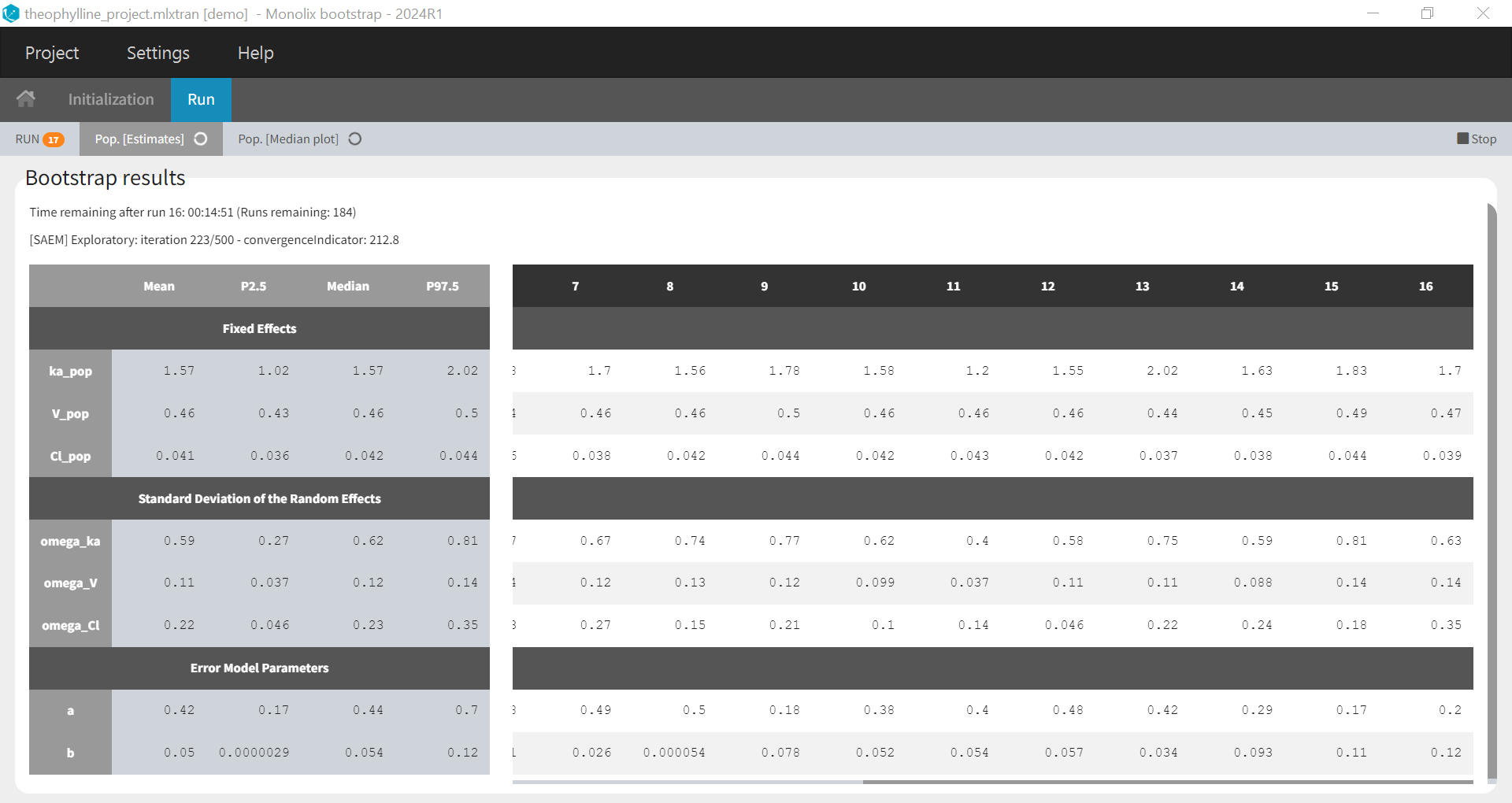
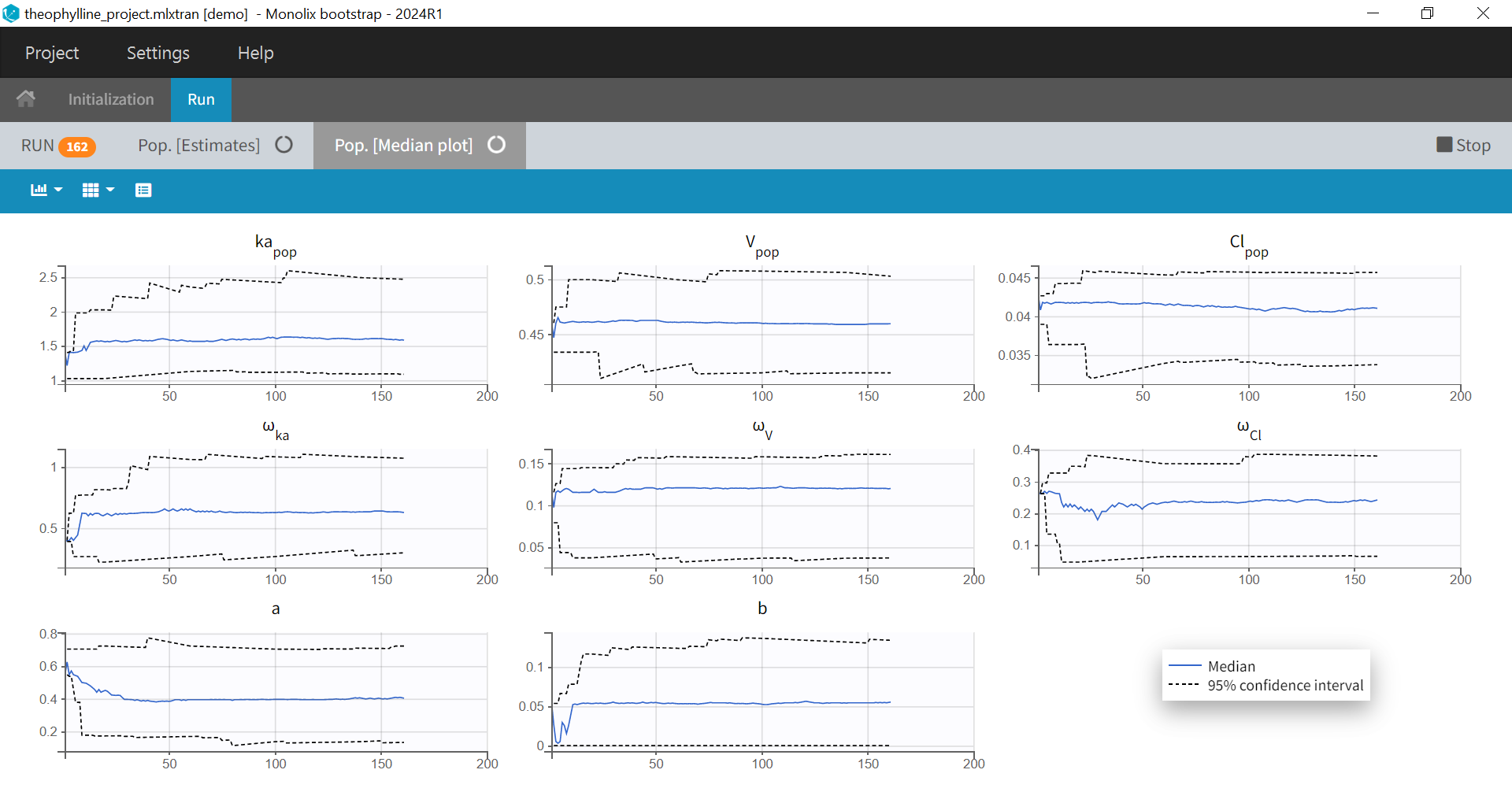
If you have stopped bootstrap before the last run, or you want to add more runs to your bootstrap results, you can resume bootstrap so that the runs already done will be reused instead of being rerun:

Results
Bootstrap results reported in the interface and the output files include the following. Depending on the choice of estimation tasks in the bootstrap settings, results for population parameters, RSE (if Standard error task was selected), and LL (if log-likelihood task was selected) are available.
-
Results > [Estimates]: Estimates from each bootstrap run displayed in tables.
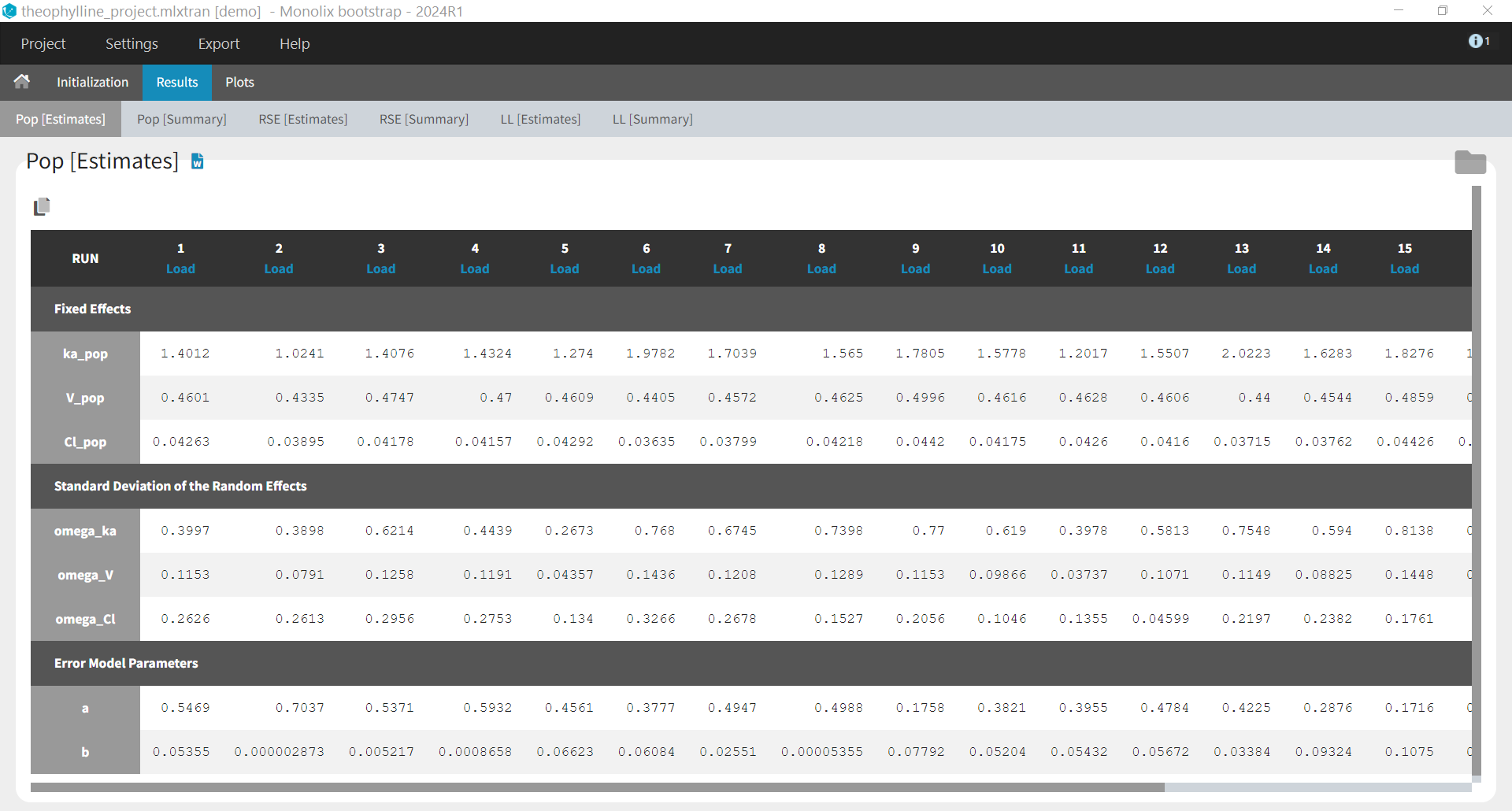
When the options to save both the results folder and the datasets have been selected, it is possible to load each bootstrap run via the “load” button in the “Pop [Estimates]” tab:
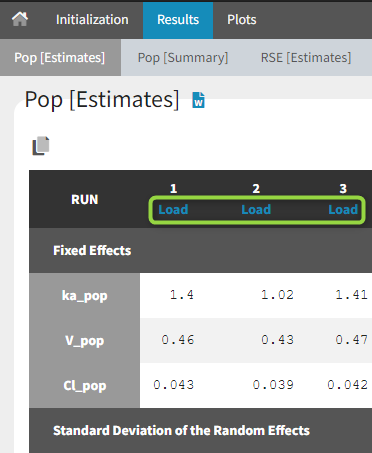
-
Plots: Estimates from each bootstrap run displayed as distribution plots. Distribution plots can be displayed as boxplots or as histograms (pdf).
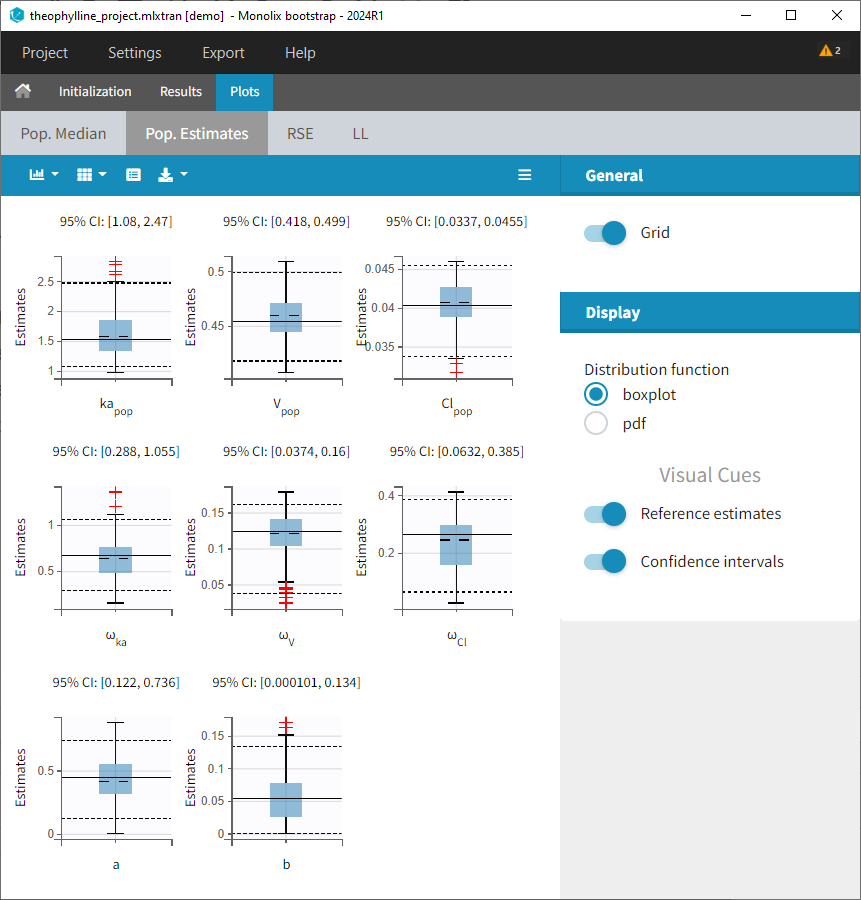
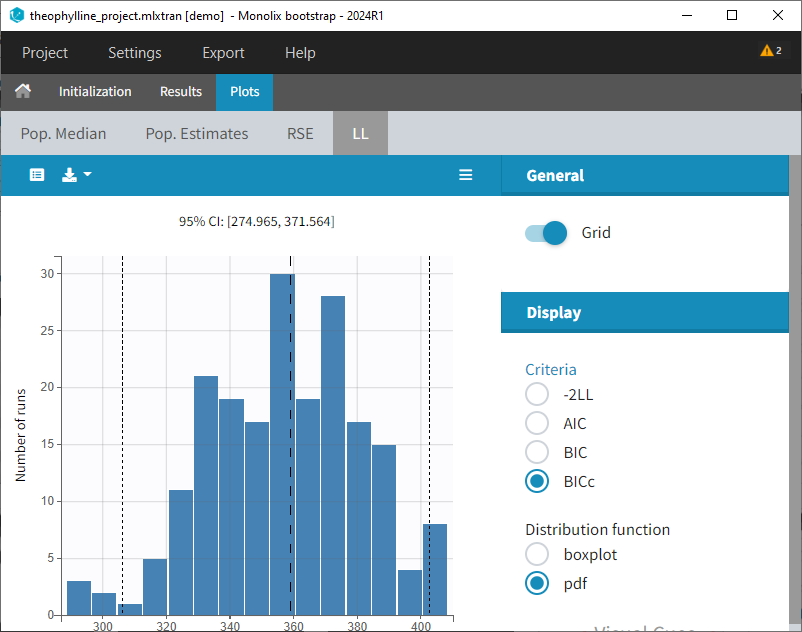
-
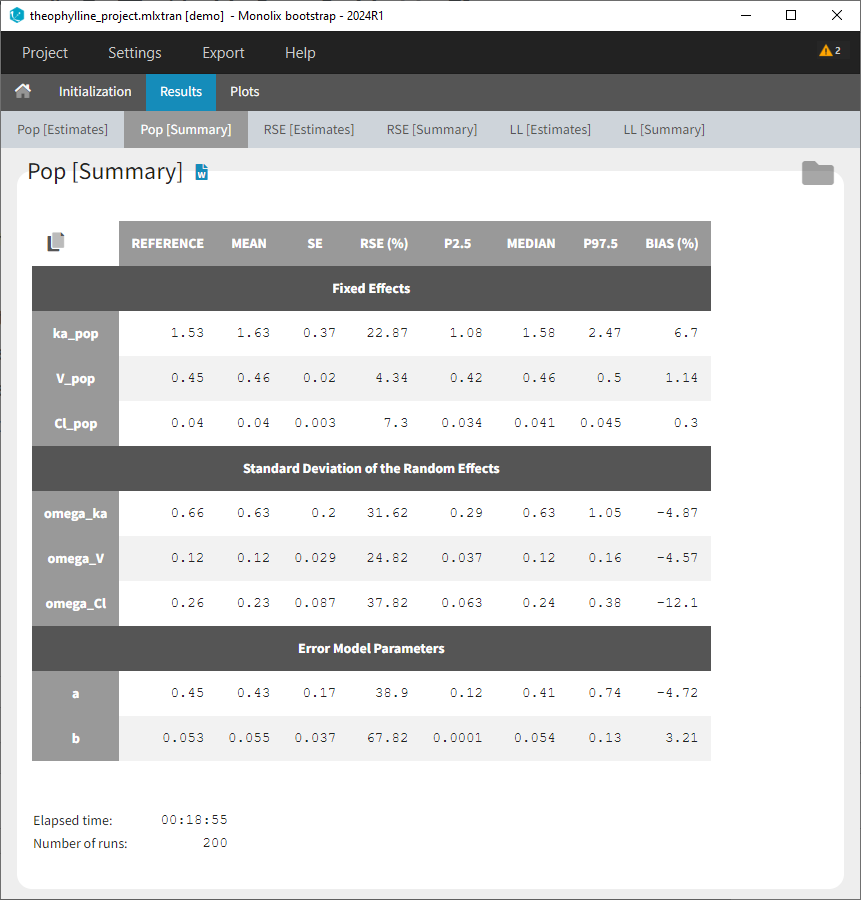
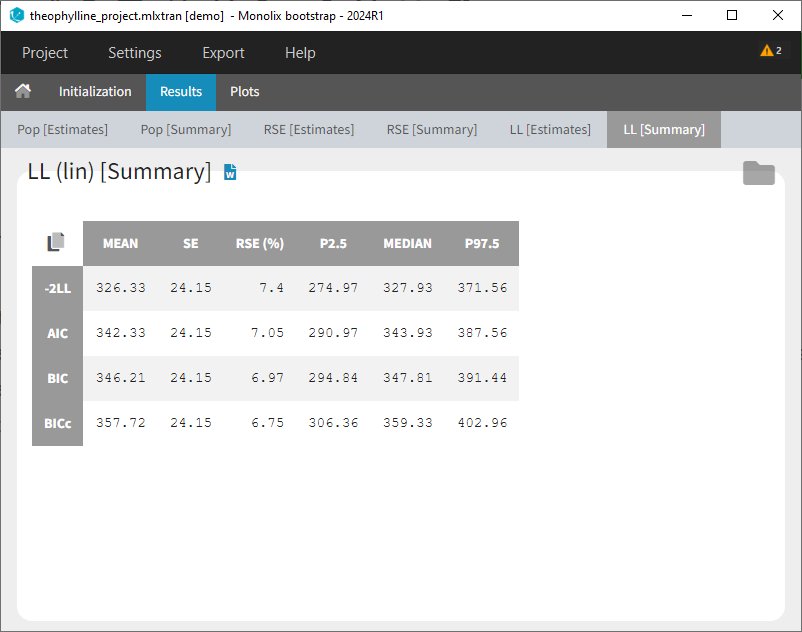
Results > [Summary]: Summary table of bootstrap estimates including value of the initial run (reference), mean and median over bootstrap runs, standard error (SE) over bootstrap runs, relative standard error (RSE) over bootstrap runs, confidence intervals (according to chosen confidence level). For population parameter, the bias of the mean over bootstrap runs compared ot the reference value is calculated as bias = (mean – reference)/reference*100.
If some runs have not reached convergences, a toggle appears to filter the summary table and keep only runs which have converged.
-
Main window (Estimation) > Results > Pop Param: The bootstrap results also appear in the table of population parameter estimates of the parent run. This is convenient to compare the estimated values and their confidence intervals estimated via the Fisher Information Matrix, to the median and confidence intervals estimated via bootstrap. Bootstrap results can be included in the pop param table when generating reports from Monolix.
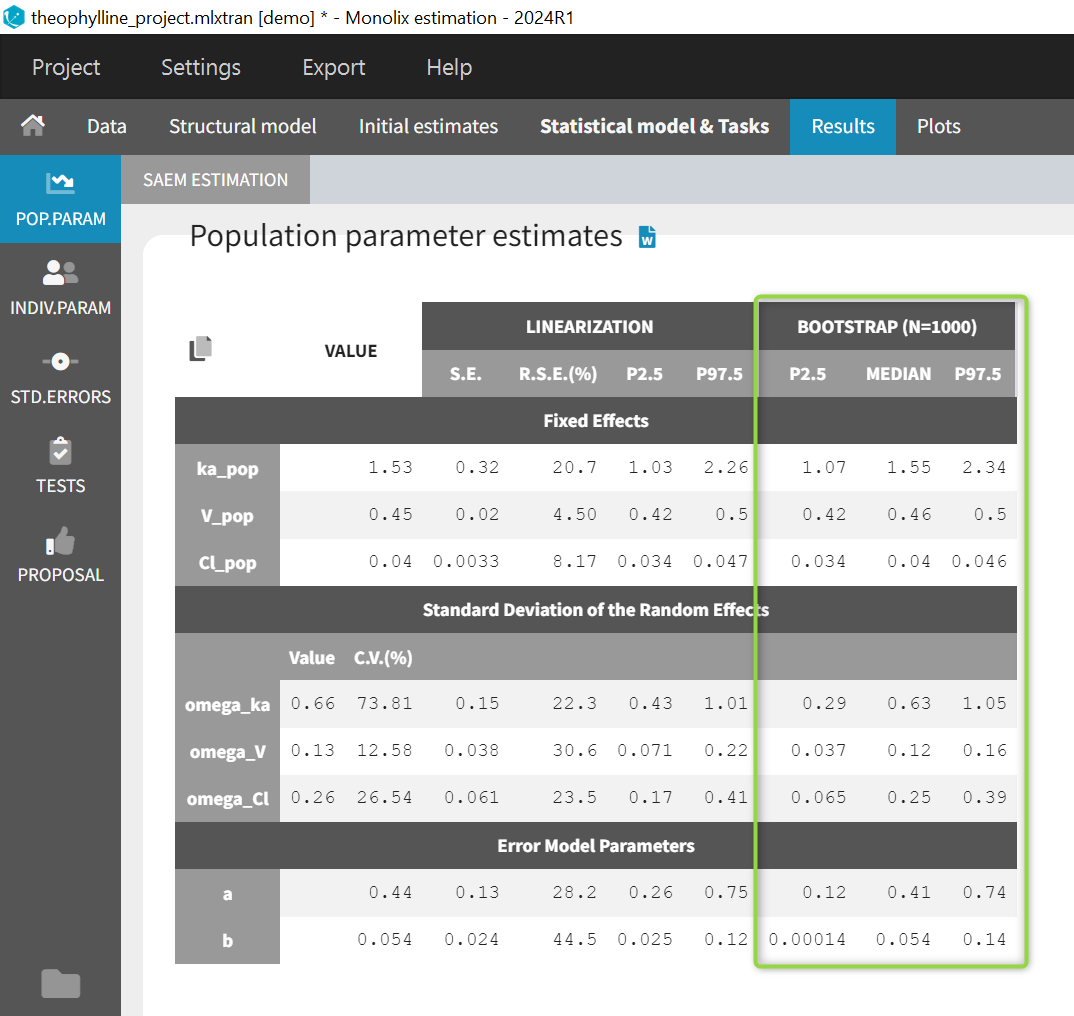
-
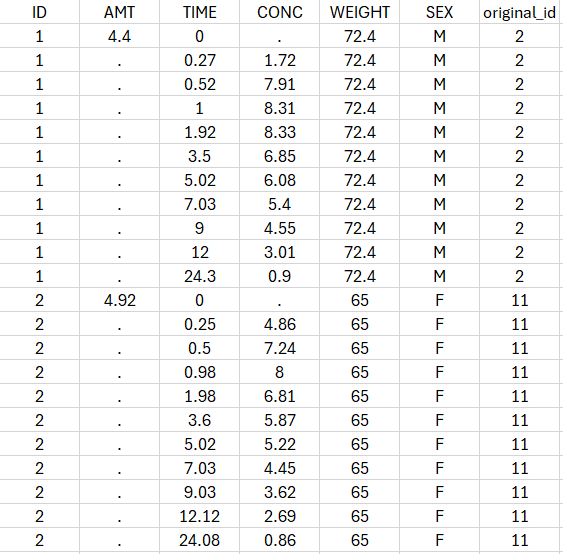
Saved datasets: In the case of non-parametric bootstrap, resampled datasets used in bootstrap runs have new subject identifiers defined as integers in the column tagged as ID, and include an additional column named “original_id” with the corresponding subject identifiers from the initial dataset.