Using or editing models from the libraries
Nine different libraries are available in Simulx, detailed in Libraries of models
PK (pharmacokinetics)
PD (pharmacodynamics)
PKPD (joint PKPD)
PK double absorption
Parent metabolite
TMDD (target-mediated drug disposition)
TTE (time to-event)
Count (for non-continuous count data)
TGI (tumor growth and tumor growth inhibition)
Picking a model from a library
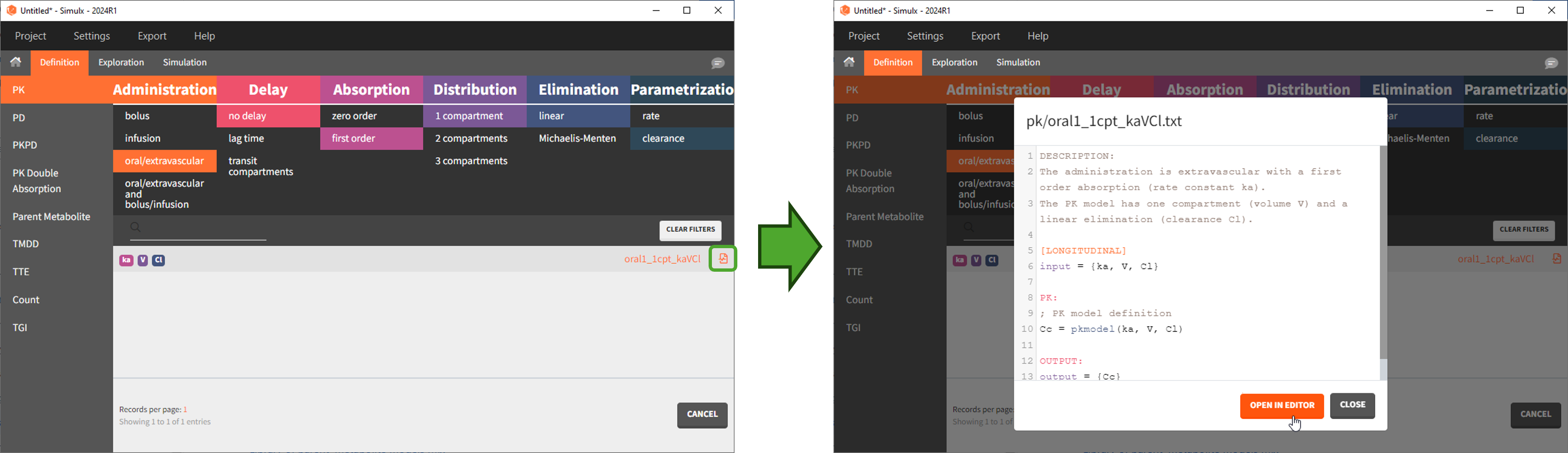
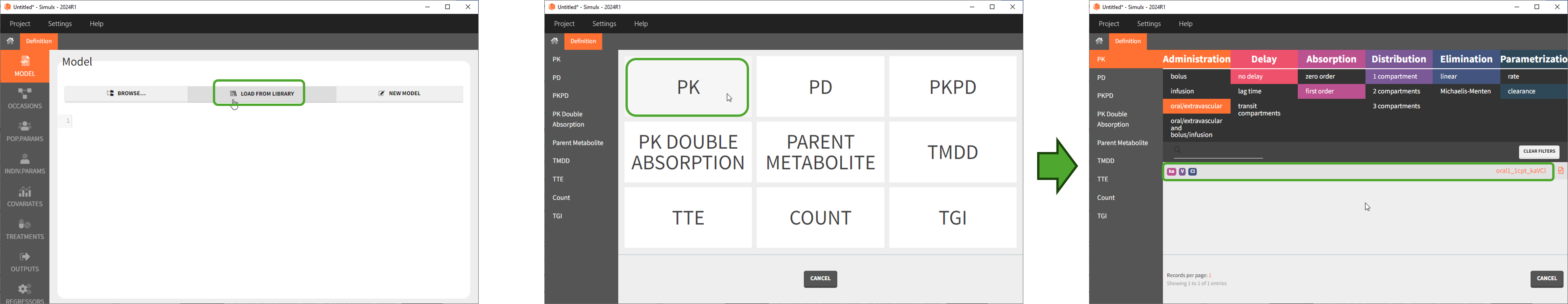
To use a model from the libraries, in the Structural model tab, click on “Load from library” and select the desired library. A list of models appears, as well as a menu to filter them. Use the filters and the list of parameters included in the models to select the model you need.
Here is for example how to select a PK model with first-order absorption and linear elimination (parameters ka, V, Cl):
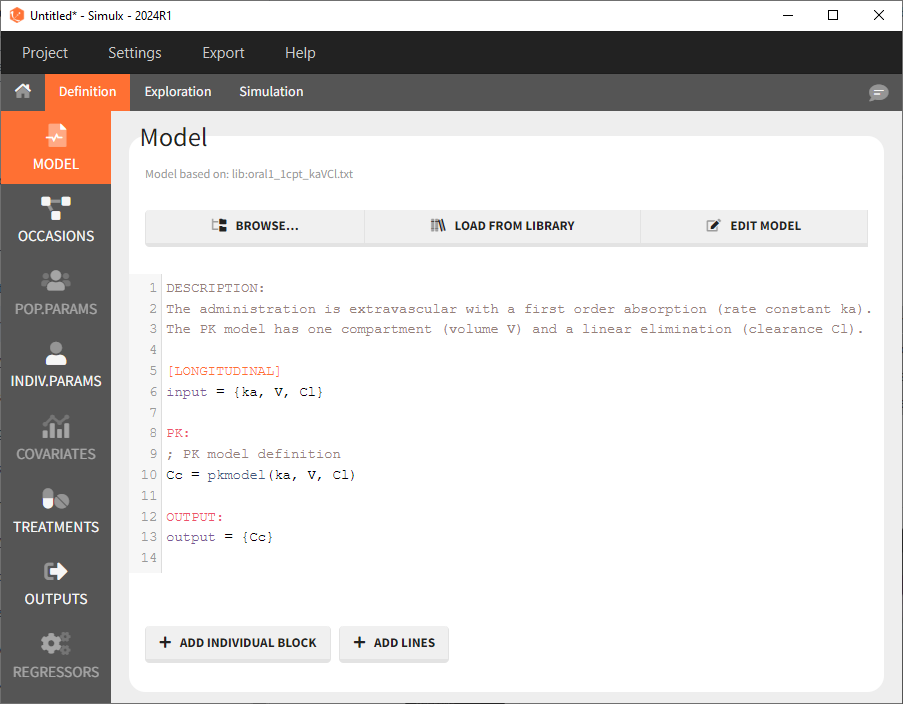
The model contents correspond to pre-written models in Mlxtran l anguage. In the case of library models, they are not saved as external text files but are generated on-the-fly inside Simulx. Once selected, the model appears in the Simulx GUI. Below we show the content of the (ka,V,Cl) model:
Modifying a model from the libraries
Browse existing models from the libraries using the “Load from library” button, then click the “file” icon next to a model name. This opens a pop-up window where the content of the model file is displayed. Click on “Open in editor” to open the model file in the MlxEditor. There you can adapt the model, for instance to add a PD model. Be careful to save the new model under a new name, to avoid overwriting the library files.