piecewise macros
Introduction
The mlxtran language provides a set of macros which allow to define a model piecewise, by defining each compartment and the transfer between compartments. The purpose of these macros is to simplify the modeling of classical pharmacokinetics. The PK model is inferred from the ensemble of used macros and the provided set of named arguments. The piecewise macros must be used within the block PK:.
An example is shown below:
PK:
; definition of the central compartment, numbered 1, with volume V and concentration Cc
compartment(cmt=1, volume=V, concentration=Cc)
; definition of an oral absorption with first-order rate ka and bioavailability F for doses with admid=1
; arriving into the central compartment. Note that the depot compartment is implicit.
absorption(adm=1, cmt=1, ka, p=F)
; definition of an iv administration into central compartment for doses with admid=2
iv(adm=2, cmt=1)
; definition of an elimination from the central compartment 1 with clearance Cl
elimination(cmt=1, Cl)Compartment macro
Purpose
The macro compartment defines a PK model compartment. Compartments can be the target of an administration, can be subject to a clearance process or can be linked with other compartments for exchange processes. The compartment amount, concentration and volume are variables.
Arguments
Arguments for the macro compartment are:
cmt: Unique identifier of the compartment. Its default value is 1.
amount: Name of variable defined as the amount within the compartment. Its dynamics are defined by dose absorptions, eliminations, and the transfer rates with other compartments.
volume: Name of predefined variable to use as the volume of the compartment. It enables to define concentration. Its default value is 1.
concentration: Name of the variable defined as the concentration within the compartment.
Basic Example
PK:
; To define a compartment with number 1, of volume V, an amount called Ac, and a concentration called Cc
compartment(cmt=1, amount=Ac, volume=V, concentration=Cc)If no administration is involved for a compartment, and if the amount and all its dynamics are defined as an ODE component with its derivative, then the compartment macro definition is not needed.
Complete Example
In the following example, a compartment is defined in the PK: bloc and the dynamics of the amount is defined with the iv and elimination macro:
[LONGITUDINAL]
input = {V, k}
PK:
compartment(cmt=1, amount=Ac, concentration=Cc, volume=V)
iv(cmt=1, type=1)
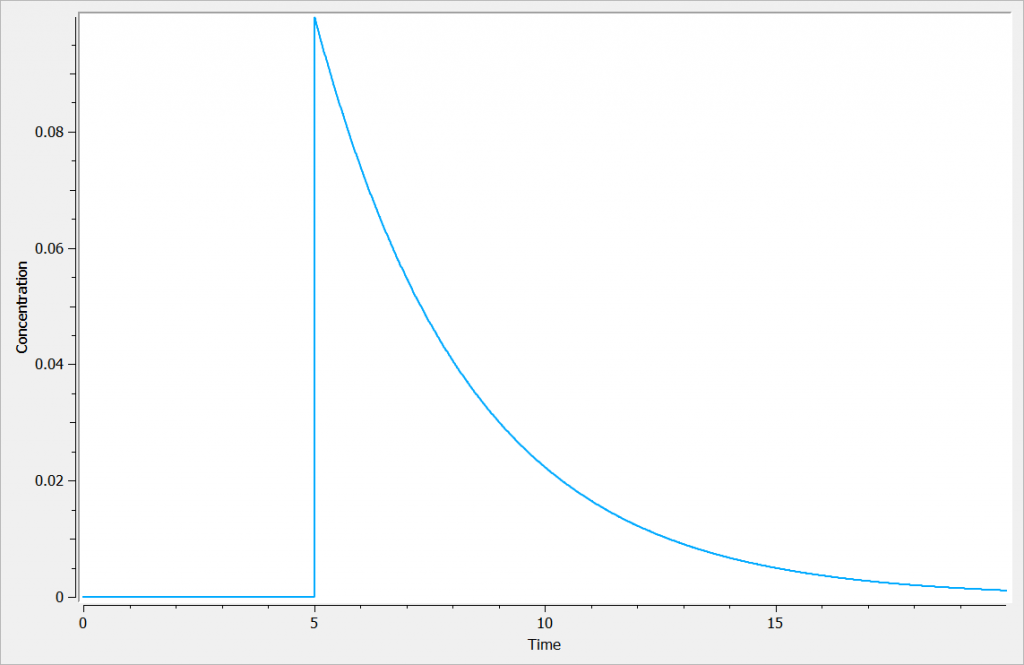
elimination(cmt=1, k)Assuming the administration of a dose of amount=1 at time=5, we obtain the following for the concentration Cc:
Rules and Best Practices
We encourage the user to use all the fields in the macro to guarantee non confusion between concentration, amount, …
CAUTION: if the volume is not given, it will be assumed to be equal to 1. The volume is in particular needed to calculate k from the clearance when Cl is used as input of the elimination macro.The compartment definition has to be done in the first place before additional macros.
Format restriction (non compliance will raise an exception)
The value after cmt= is necessarily an integer.
The value after amount=, volume= and concentration= are necessarily strings.
Peripheral macro
Purpose
The peripheral macro defines a peripheral compartment. It is equivalent to a compartment with two transfers of drug amount towards and from another base compartment. This base compartment must have been previously defined and referenced by the indexes used in the parameter names: peripheral(k12, k21) defines a peripheral compartment numbered 2, which will be connected to the already defined compartment 1. If the amount or concentration of the peripheral compartment are not used afterwards, then they do not need to be defined as part of the macro.
Arguments
Arguments for macro peripheral are:
kij or ki_j: Input rate from the compartment of label i. It also defines a label j for the peripheral compartment. Here, both labels i and j must be integers. If i and/or j are larger than 9, the syntax with the underscore is necessary. Mandatory.
kji or kj_i: Output rate to the compartment of label i. It also defines a label j for the peripheral compartment. Here, both labels i and j must be integers. If i and/or j are larger than 9, the syntax with the underscore is necessary. Mandatory.
amount: Name of variable defined as the amount within the compartment. Its dynamics are defined by dose absorptions, eliminations, and the transfer rates with other compartments.
volume: Name of predefined variable to use as the volume of the compartment. It enables to define concentration. The default value is 1. If the volume is not defined, the concentration cannot be defined.
concentration: Name of the variable defined as the concentration within the peripheral compartment.
Most of the time it is not necessary to name the amount, volume and concentration.
Example
PK:
; definition of a peripheral compartment with cmt=2 with rates k12 and k21
peripheral(k12, k21)
; if the concentration in the peripheral compartment is measured
; a volume V2, and a concentration Cp can also be defined
peripheral(k12, k21, volume=V2, concentration=Cp)
; definition of a peripheral compartment with cmt=3, linked to compartment 1,
peripheral(k13, k31)
; using Q and V2 instead
peripheral(k12=Q/V, k21=Q/V2); with compartments numbers larger than 9
peripheral(k2_13, k13_2)Rules and best practices
To use inter-compartment clearance Q and peripheral volume V2 as parameters instead of k12 and k21, the syntax is:
CODEperipheral(k12=Q/V, k21=Q/V2)Format restriction (non compliance will raise an exception)
The values after k are necessarily an integer (related to predefined compartment id).
The value after kij= can be either a double or replaced by an input parameter.
The value after amount= and concentration= are necessarily strings.
The value after volume= can be either a double or replaced by an input parameter.
Effect macro
Purpose
The macro effect defines an effect compartment. The effect compartment is usually linked to a central compartment. The drug exchange between the effect compartment and the central compartment does not affect the mass balance of the amount in the central compartment (its impact is considered negligible). The central compartment must be defined before the effect compartment is defined.
Arguments
Arguments for the macro effect are:
cmt: Label of the linked base compartment. Its default value is 1.
ke0: Transfer rate from the linked base compartment. Mandatory.
concentration: Name of the variable defined as the concentration within the effect compartment. Mandatory.
Basic Example
PK:
; Define an effect compartment linked to the base compartment 1,
; with a transfer rate ke0 to the effect compartment,
; with Ce as concentration's name
effect(cmt=1, ke0, concentration=Ce)Complete Example:
In the following example, a compartment is defined in the PK: bloc. An iv absorption process along with a linear elimination process is added to the main compartment. An effect compartment is added with a transfer rate ke0.
[LONGITUDINAL]
input = {V, k, ke0}
PK:
compartment(cmt=1, amount=Ac, concentration=Cc, volume=V)
elimination(cmt=1, k)
iv(cmt=1)
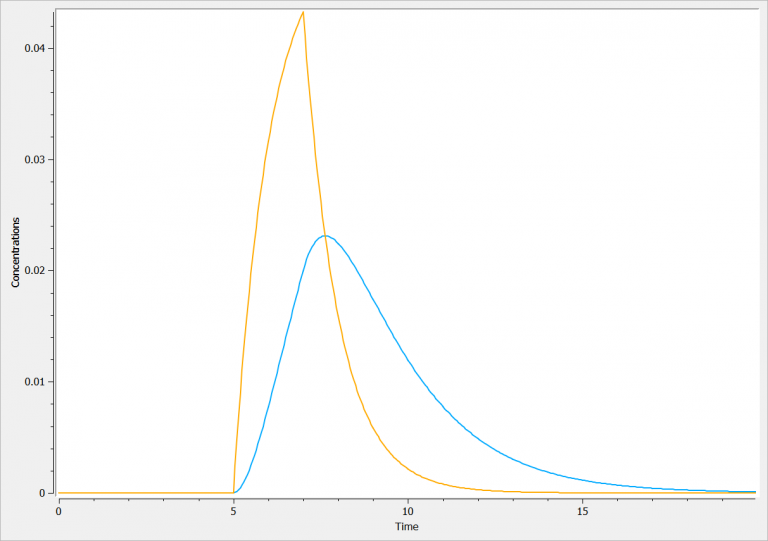
effect(cmt=1, ke0, concentration=Ce)Assuming a single infusion at time=5 for a duration of 2hr, the concentration in the central compartment (orage) and in the effect compartment (blue) are:
Rules and Best Practices
We encourage the user to use all the fields in the macro to guarantee non confusion between fields.
A base compartment must be defined first.
Format restriction (non compliance will raise an exception)
The value after cmt= is necessarily an integer.
The value after ke0= can be either a double or replaced by an input parameter. Calculations are not supported.
The value after concentration= is necessarily a string.
Transfer macro
Purpose
The macro transfer defines a uni-directional transfer process from a base compartment to a target compartment. The base compartment and the target compartment must be defined first before the transfer can be defined. For the more common bi-directional transfer process it is better to use the macro peripheral instead of the macro transfer, in particular as it allows to use the analytical solution, which is not the case with the macro transfer.
Arguments
Its named arguments are:
from: Label of the source compartment for the transfer. Its default value is 1.
to: Label of the target compartment for the transfer. Its default value is 1.
kt: Rate of the transfer. Mandatory.
Basic Example
PK:
; transfer from compartment 1 to compartment 2 with a rate kt
transfer(from=1, to=2, kt)
Complete Example
In the following example, a sigmoid absorption model is sown. Two compartments are defined in the PK block to specify a depot and a central compartment. The oral() macro define a zero-order absorption into the depot compartment (A transfer from compartment 1 to compartment 2 is added, with a transfer rate kt.
<MODEL>
[LONGITUDINAL]
input = {Tk0, ka, V, Cl}
PK:
; depot compartment
compartment(cmt=1, amount=Ad)
; central compartment
compartment(cmt=2, concentration=Cc, volume=V)
; zero-order absorption into the depot compartment 1
oral(cmt=1, Tk0)
; first-order transfer from depot (1) to central (2)
transfer(from=1, to=2, kt=ka)
; elimination from central compartment (2)
elimination(cmt=2, Cl)Best Practices
We encourage the user to use all the fields in the macro to guarantee non confusion between the fields.
Format restriction (non compliance will raise an exception)
The value after from= and to= are necessarily integers.
The value after kt= can be either a double or replaced by an input parameter. Calculations are not supported.
Absorption/oral macro
Purpose
The macro absorption/oral enables to link the administration and the model, for zero-order and first-order absorption processes (for bolus administrations, use the iv macro instead). For Mlxtran the macro names ‘absorption’ and ‘oral’ can be used interchangeably. The administration can either be defined in a data set when Monolix is used, or the administration can be defined via an administration design definition when Simulx is used. In order to handle models with several administrations each administration has a unique identifier called “adm”. This identifier must be defined in the data set or in the administration design definition, and used in the absorption/oral macro.
Arguments common to zero-order and first-order
The absorption/oral macro can be used for zero-order and first-order absorptions. The arguments common to the two absorptions are:
adm: Administration identifier of doses to be read by this macro. Its default value is 1. Alias: type.
Tlag: Lag time before the absorption. Its default value is 0.
p: Final proportion of the absorbed amount. It can affect the effective rate of the absorption, not its duration. p can take any positive value (including > 1). Its default value is 1.
cmt: Label of the compartment called in by the absorption process. Its default value is 1. The corresponding compartment must be defined before using the absorption/oral macro using a compartment macro.
To define a zero-order or first-order absorption, the additional arguments defined below are needed.
Arguments specific to a zero-order absorption
To define a zero-order absorption, the following argument must be added:
Tk0: Duration of the zero order absorption. Mandatory to define a zero-order absorption.
Example:
; zero order absorption process for the doses of type 1, in compartment 1 with a delay Tlag of 1 and a duration Tk0
absorption(adm=1, cmt=1, Tlag=1, Tk0 = 2, p=1)Arguments specific to a first-order absorption
To define a first-order absorption, the following arguments can be added:
ka: Rate of the first order absorption. Mandatory to define a first-order absorption.
Ktr: Transit rate.
Mtt: Mean transit time for the absorption.
Example:
PK:
; first order absorption process for the doses of type 1, in compartment 1 with a delay Tlag of 1 and a rate ka
absorption(adm=1, cmt=1, Tlag=1, ka)
Note: in case of transit compartments, the mean transit time is defined as Mtt = (n+1)/Ktr with n the number of transit compartments (excluding the depot compartment and the absorption compartment):

The implementation is the same as described in Savic et al, “Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies” (2007), except that to approximate the factorial n!, we use the gamma function, which is more precise, instead of the Stirling formula.
In case of multiple doses, we assume that the previous dose has been completely absorbed (i.e concentration is the Abs compartment is almost zero) when the next dose is administered. If this is not the case, the remaining concentration in the Abs compartment is erased when the next dose is applied, leading to a loss of dose amount entering the system.
Example with mixed absorptions
Some drugs can exhibit atypical absorption profiles with for instance a zero-order absorption followed by a first-order absorption, or first and zero-order simultaneously.
Sequential absorption, zero-order followed by first-order
In the example below, a fraction F1 of the drug is absorbed via a zero-order process for a duration Tk0. The remaining fraction 1-F1 is then absorbed via a first-order process, which starts with a lag-time Tk0 (i.e at the end of the zero-order absorption phase).
PK:
absorption(adm=1, cmt=1, Tk0, p=F1)
absorption(adm=1, cmt=1, ka, p=1-F1, Tlag=Tk0)Simultaneous first-order and zero-order absorption
In the example below, a fraction F1 of the drug is absorbed via a first-order process (with absorption rate ka). Simultaneously the remaining fraction 1-F1 is absorbed via a zero-order process.
PK:
absorption(adm=1, cmt=1, ka, p=F1)
absorption(adm=1, cmt=1, Tk0, p=1-F1)A lag time for one or the other process can be added if necessary.
Example with different absorption models
In the presented example, we show the difference between the two absorption processes. In addition, the effect of the Ktr and Mtt parameters is explored in a third model.
[LONGITUDINAL]
input = {V, Tlag, ka, Tk0, Ktr, Mtt}
PK:
; model 1 with zero-order absoprtion
compartment(cmt=1, amount=A1, concentration=Cc_zoa, volume=V)
absorption(cmt=1, adm=1, Tk0)
elimination(cmt=1, k=1)
; model 2 with first-order absorption
compartment(cmt=2, amount=A2, concentration=Cc_foa, volume=V)
absorption(cmt=2, adm=1, Tlag, ka)
elimination(cmt=2, k=1)
; model 3 with transit compartments
compartment(cmt=3, amount=A3, concentration=Cc_foaT, volume=V)
absorption(cmt=3, adm=1, Tlag, ka, Ktr, Mtt)
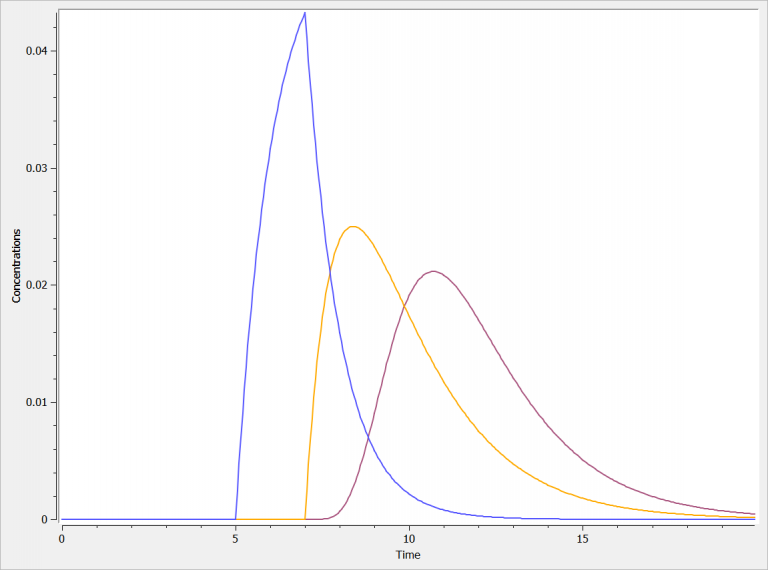
elimination(cmt=3, k=1)The three concentrations are presented in the following figure:
zero order absorption (blue): the peak concentration is sharp
first order absorption (orange): the peak concentration is more smooth. In addition, is this example it is delayed before of the Tlag
transit compartments (purple): the start of the absorption is delayed and progressive
Rules and Best Practices
Format restriction (non compliance will raise an exception)
The value after cmt= and type= are necessarily integers.
The value after Tlag=, p=, V=, ka=, Tk0=, Ktr=, Mtt= can be either a double or replace by an input parameter. Calculations are not supported.
The associated treatment or administration can not be defined with a rate nor an infusion timing (RATE and TINF column-types in the data set). If a rate or an infusion timing is present, the user can use the iv macro and/or define a compartment to define the dynamics of the absorption.
IV macro
Purpose
The macro iv enables to link the administration with the model, for intravenous doses (bolus or infusion). Doses defined in the data set without an administration rate or infusion time are instantaneously absorbed within the associated compartment, as an IV bolus. Doses with an administration rate or infusion time are absorbed according to a zero order process, as an IV infusion.
Arguments
Arguments for macro iv() are:
cmt: Label of the compartment called in by the absorption process. Its default value is 1. The corresponding compartment must be defined before using the absorption/oral macro using a compartment macro.
adm: Administration identifier of doses to take in. Its default value is 1. Alias: type.
Tlag: Lag time before the absorption. Its default value is 0.
p: Final proportion of the absorbed amount. It can affect the effective rate of the absorption, not its duration. p can take any positive value (including > 1). Its default value is 1.
Basic Example
PK; intravenous bolus for the doses of type 1, in compartment 1 with a delay Tlag at 1
iv(adm=1, cmt=1, Tlag=1, p=1)Complete Example
In the presented example, we show the difference two iv processes with and without a rate in the administration.
[LONGITUDINAL]
input = {V, k}
PK:
compartment(cmt=1, concentration=Cc, volume=V)
iv(cmt=1, adm=1)
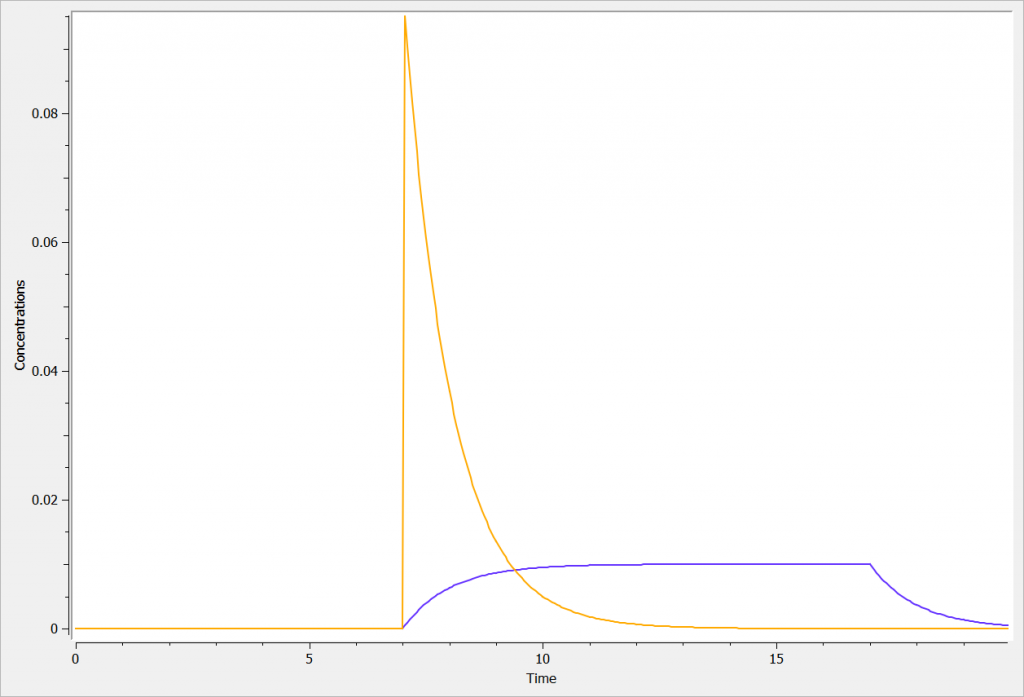
elimination(cmt=1, k)The concentration in yellow shows the profile after an iv bolus dose. The concentration in blue shows the profile after an iv infusion of 17 minutes.
Rules and Best Practices
Format restriction (non compliance will raise an exception)
The value after cmt= and adm= are necessarily integers.
The value after Tlag=, p= can be either a double or replaced by an input parameter. Calculations are not supported.
With the iv macro, it is not possible to give the rate or duration of the infusion as a parameter or fixed value. To do so, use the absorption macro with duration parameter Tk0 to define a zero-order absorption (i.e infusion).
When the iv macro is used in combination with a data set with a column-type RATE:
if RATE >0: infusion with rate RATE and duration AMT/RATE
if RATE <=0: bolus
When the iv macro is used in combination with a data set with a column-type TINF:
if TINF >0: infusion of duration TINF at a rate AMT/TINF
if TINF <=0: bolus
Empty and reset macros
Description
The macro empty can be used to set any component of the system, defined in an ODE, to 0 at any given time. Similarly, the macro reset is used to reset a variable of the system to its initial value. Empty or reset times are indicated via a type of administration (adm id). The actions of these macros are thus selective, contrary to the reset applied with an EVID column in the dataset with values 3, which resets all compartments.
Arguments
The arguments are the same for both empty and reset macros:
adm: Administration identifier used to indicate empty or reset times. Its default value is 1. Alias: type. The empty and reset macrso only use the administration times, not the amounts.
target: Name of the ODE variable that is set to 0 or reset to its initial value at the specified times, or target=all to reset all system variables. Mandatory.
Basic Example
The following code defines an emptying of the ODE variable Ap on administration times of type 1 (e.g ADM=1 in the data set).
PK:
empty(adm=1, target=Ap)This is equivalent to the following code with the depot macro:
PK:
depot(adm=1, target=Ap, p=-Ap/amtDose)The following code defines a reset based on administration times of type 2 (e.g ADM=2 in the data set), applied to the ODE variable Ac.
PK:
reset(adm=2, target=Ac)It is possible to define several empty macros with the same adm identifiers to set several targets to 0 at the same times, and the same for reset macros. With the example below, dose events with adm=3 will empty both variables Ac and Ap.
PK:
empty(adm=3, target=Ac)
empty(adm=3, target=Ap)To reset all variables of the system, “target=all” can al be used:
PK:
reset(adm=3, target=all)Complete example: urine compartment
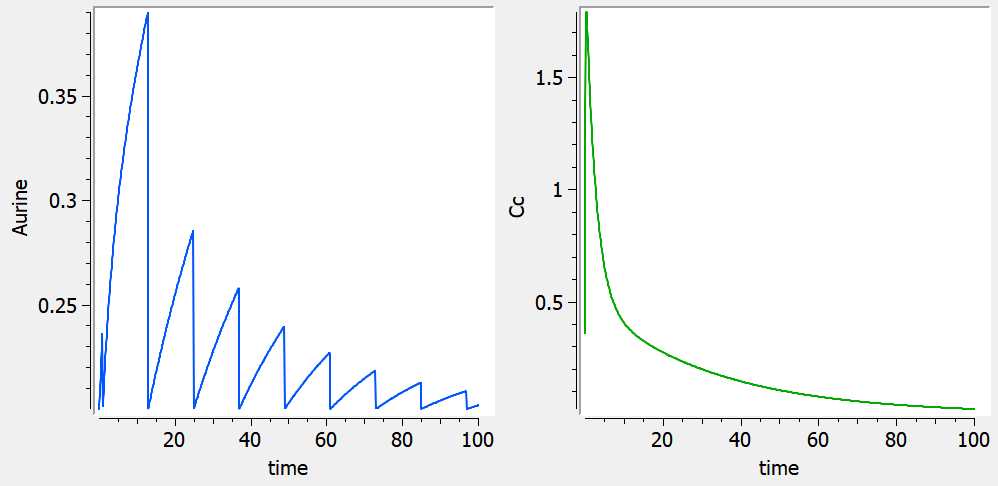
The following script implements a two compartments model with an additional urine compartment. There is a transfer from the central compartment to the urine compartment. A single dose with adm=1 is administered to the central compartment at time 0. A selective reset of the urine compartment corresponding to collecting times is defined with an administration with adm=2. Each amount for this administration is defined as 1 but is actually not used in the model.
When using this model with a data set, urine would be collected over consecutive periods of 12 hours and the amount of drug measured in the urine volume collected for each period would be recorded.
[LONGITUDINAL]
input = {ka, Cl, V1, Q, V2, p_urine}
PK:
k_urine = p_urine*Cl/V1
k_non_urine = (1-p_urine)*Cl/V1
k12 = Q/V1
k21 = Q/V2
; Dose administration to central compartment (plasma)
depot(adm=1, target=Ac, ka)
; Reset of urine compartment
reset(adm=2, target=Aurine)
EQUATION:
t_0=0
Ac_0=0
Ap_0=0
Aurine_0=0
ddt_Ac = - k_non_urine*Ac - k12*Ac + k21*Ap - k_urine*Ac
ddt_Ap = k12*Ac-k21*Ap
ddt_Aurine = k_urine*Ac
Cc=Ac/V1
OUTPUT:
output = {Cc, Aurine}The model gives the following profiles for the drug amount in the urine compartment (blue, left), and the drug concentration in the central compartment (green, right).
Complete example: PD variable
Emptying a compartment or resetting it to its initial state are equivalent actions for compartment which are initially empty. This may be not the case for PD models with non-zero baseline, like in the following example where the PD variable E is reset to its initial value E0 at time 60 and 108 using a dummy dose with ADMID=2.
<MODEL>
[LONGITUDINAL]
input = {ka, k, E0, IC50, kout}
PK:
depot(target=Ad, type=1)
reset(target=E, type=2)
EQUATION:
E_0 = E0
kin = E0*kout
ddt_Ad = -ka*Ad
ddt_Ac = ka*Ad - k*Ac
ddt_E = kin*(1 - Ac/(Ac+IC50)) - kout*E
OUTPUT:
output = {E}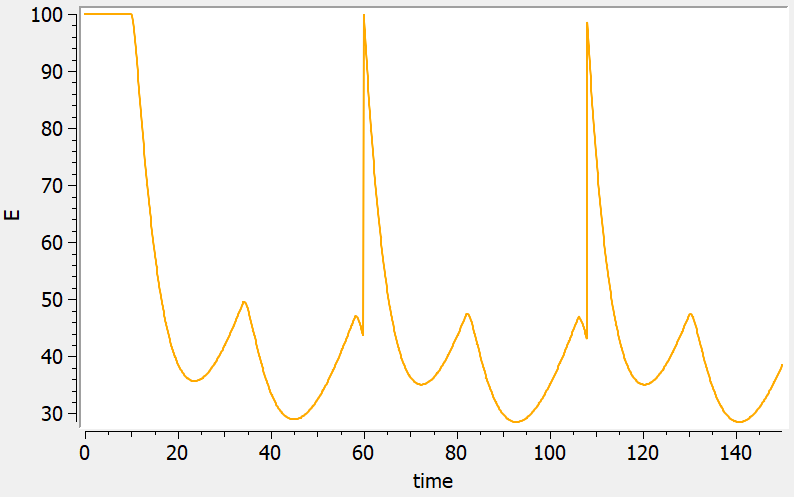
Rules
The value after type= or adm= must be an integer.
The value after target= must be a string representing the ODE variable.
Elimination macro
Purpose
The elimination macros permits to define different elimination processes (linear or Michaelis-Menten) for compartments. Several eliminations can be defined for the same compartment.
Arguments
Some arguments are common to the two elimination types, while some are specific to a linear or to a Michaelis-Menten type of elimination.
Arguments common to linear and Michaelis-Menten:
cmt: Label of the compartment emptied by the elimination process. Its default value is 1.
Arguments specific to a linear elimination
The use of one of the following additional arguments defines a linear elimination:
k: Rate of the elimination.
Cl: Clearance of the elimination.
Only one of k or Cl must be defined.
Note, that if no volume has been defined for the compartment, then the default volume V=1 is assumed.
Example:
PK:
elimination(cmt=1,k)Arguments specific to a Michaelis-Menten elimination
The use of both of the following additional arguments defines a Michaelis-Menten elimination:
Vm: Maximum elimination rate. The unit of Vm is amount/time.
Km: Michaelis-Menten constant. The unit of Km is a concentration.
Both arguments must be defined.
Example:
PK:
elimination(cmt=1, Vm, Km)Complete Example
The following model implemented with PK macros includes two compartments, an oral absorption and a dual elimination pathway with parallel linear and Michaelis-Menten eliminations. An equivalent model, implemented with ODEs, is defined in the TMDD model library: it corresponds to the Michaelis-Menten approximation of a TMDD model.
[LONGITUDINAL]
input = {ka, V, Vm, Km, Cl, Q, V2}
PK:
kel = Cl/V
k12 = Q/V
k21 = Q/V2
compartment(cmt=1, concentration=Cc, volume=V)
absorption(cmt=1, ka)
peripheral(k12, k21)
elimination(cmt=1, k=kel)
elimination(cmt=1, Vm, Km)
OUTPUT:
output={Cc}Rules and Best Practices
Format restriction (non compliance will raise an exception)
The value after cmt= is necessarily an integer.
The value after V=, k=, Cl=, Km=, Vm= can be either a double or be replaced by an input parameter. Calculations are not supported.
