plotStandardizedRandomEffectsDistribution
[Monolix] Distribution of the standardized random effects.
Plot the distribution of the standardized random effects.
Usage
plotStandardizedRandomEffectsDistribution(
parameters = NULL,
plot = "boxplot",
settings = list(),
preferences = list(),
stratify = list()
)Arguments
- parameters
vector of parameters to display. (by default the first 4 computed parameters are displayed).
- plot
Type of plot: probability density distribution ("pdf"), cumulative density distribution ("cdf"), boxplot ("boxplot") (default "boxplot").
- settings
a list of optional plot settings:
indivEstimateCalculation of individual estimates: conditional mean ("mean"), conditional mode with EBE's ("mode"), conditional distribution ("simulated") (default "mode").variabilityLevel(logical) In case of IOV and if the conditional distribution is computedplot is displayed for one given level of variability (default NULL) If NULL, the variability level is ID + Occasions Run getVariabilityLevels() to see the available levels of variabilityempirical(logical) If TRUE, plot empirical density distribution(default TRUE). To define when plot is "pdf" or "cdf"theoretical(logical) If TRUE, plot theoretical density distribution(default TRUE). To define when plot is "pdf" or "cdf"median(logical) If TRUE, add median line (default TRUE). To define when plot is "boxplot"quartile(logical) If TRUE, add quartile line (default TRUE). To define when plot is "boxplot"legend(logical) add (TRUE) / remove (FALSE) plot legend (default FALSE).grid(logical) add (TRUE) / remove (FALSE) plot grid (default TRUE).ncol(integer) number of columns when facet = TRUE (default 4).fontsize(integer) Plot text font size.
- preferences
(optional) preferences for plot display, run getPlotPreferences("plotStandardizedRandomEffectsDistribution") to check available displays.
- stratify
List with the stratification arguments:
groups- Definition of stratification groups. By default, stratification groups are already defined as one group for each category for categorical covariates, and two groups of equal number of individuals for continuous covariates. To redefine groups, for each covariate to redefine, specify a list with:name character covariate name (e.g "AGE")definition (vector(continuous) || list>(categorical)) For continuous covariates, vector of break values (e.g c(35, 65)). For categorical covariates, groups of categories as a list of vectors(e.glist(c("study101"), c("study201","study202")))split(vector) - Vector of covariates used to split (i.e facet) the plot (by default no split is applied). For instancec("FORM","AGE").filter(list< list> >) - List of pairs containing a covariate name and the vector of indexes or categories (for categorical covariates) of the groups to keep (by default no filtering is applied). For instance,list("AGE",c(1,3))to keep the individuals belonging to the first and third age group, according to the definition ingroups. For instance,list("FORM","ref")using the category name for categorical covariates.
Value
A ggplot object if one parameter,
A TableGrob object if multiple plots (output of grid.arrange)
See also
Examples
initializeLixoftConnectors(software = "monolix")
project <- file.path(getDemoPath(), "1.creating_and_using_models",
"1.1.libraries_of_models", "theophylline_project.mlxtran")
loadProject(project)
runPopulationParameterEstimation()
runConditionalDistributionSampling()
runConditionalModeEstimation()
# Random effect distribution as boxplot
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "boxplot")

plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "boxplot",
settings = list(indivEstimate = "mode"))

plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "boxplot",
settings = list(quartile = F))

# Random effect distribution as pdf
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "pdf")
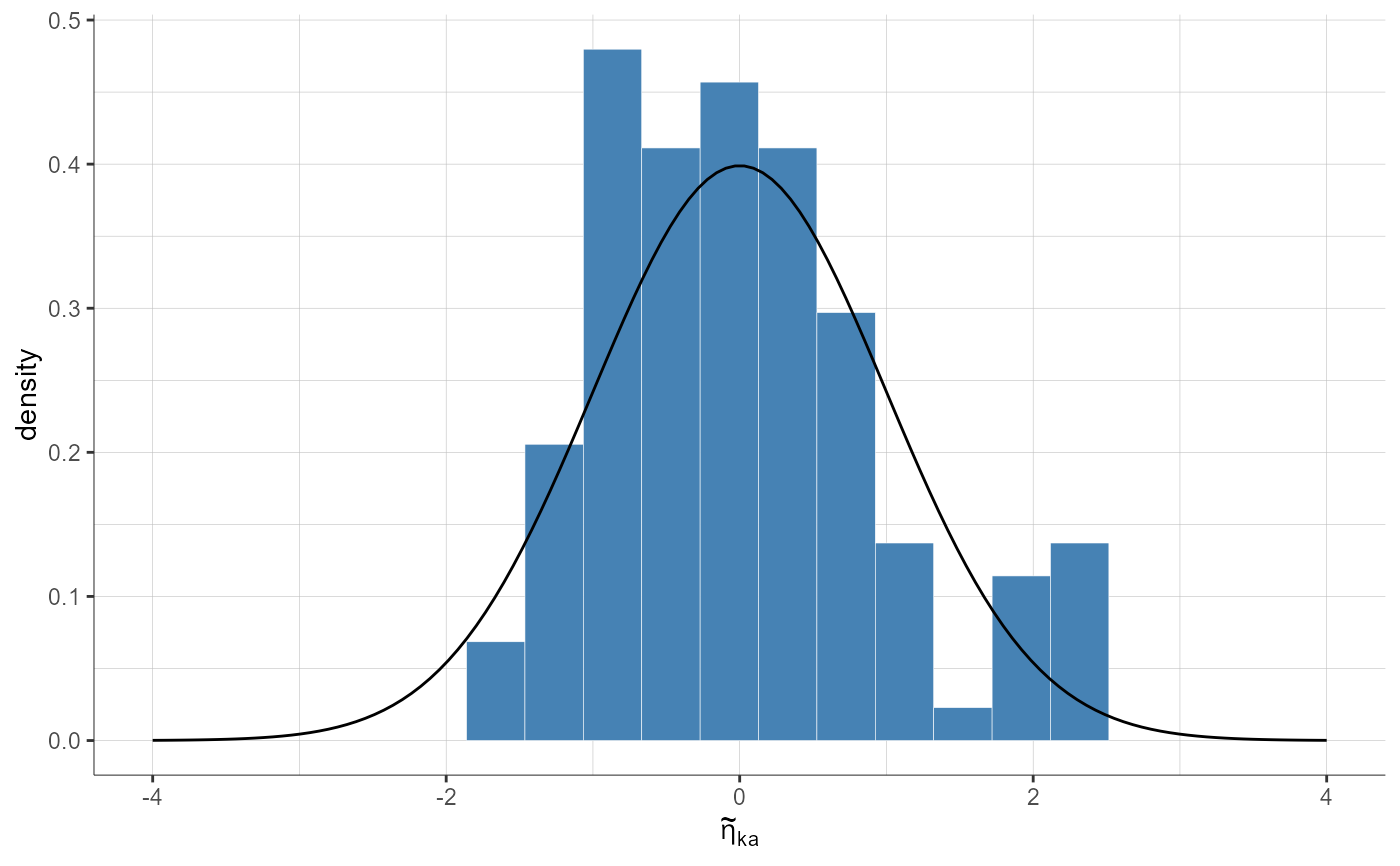
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "pdf",
settings = list(empirical = F))
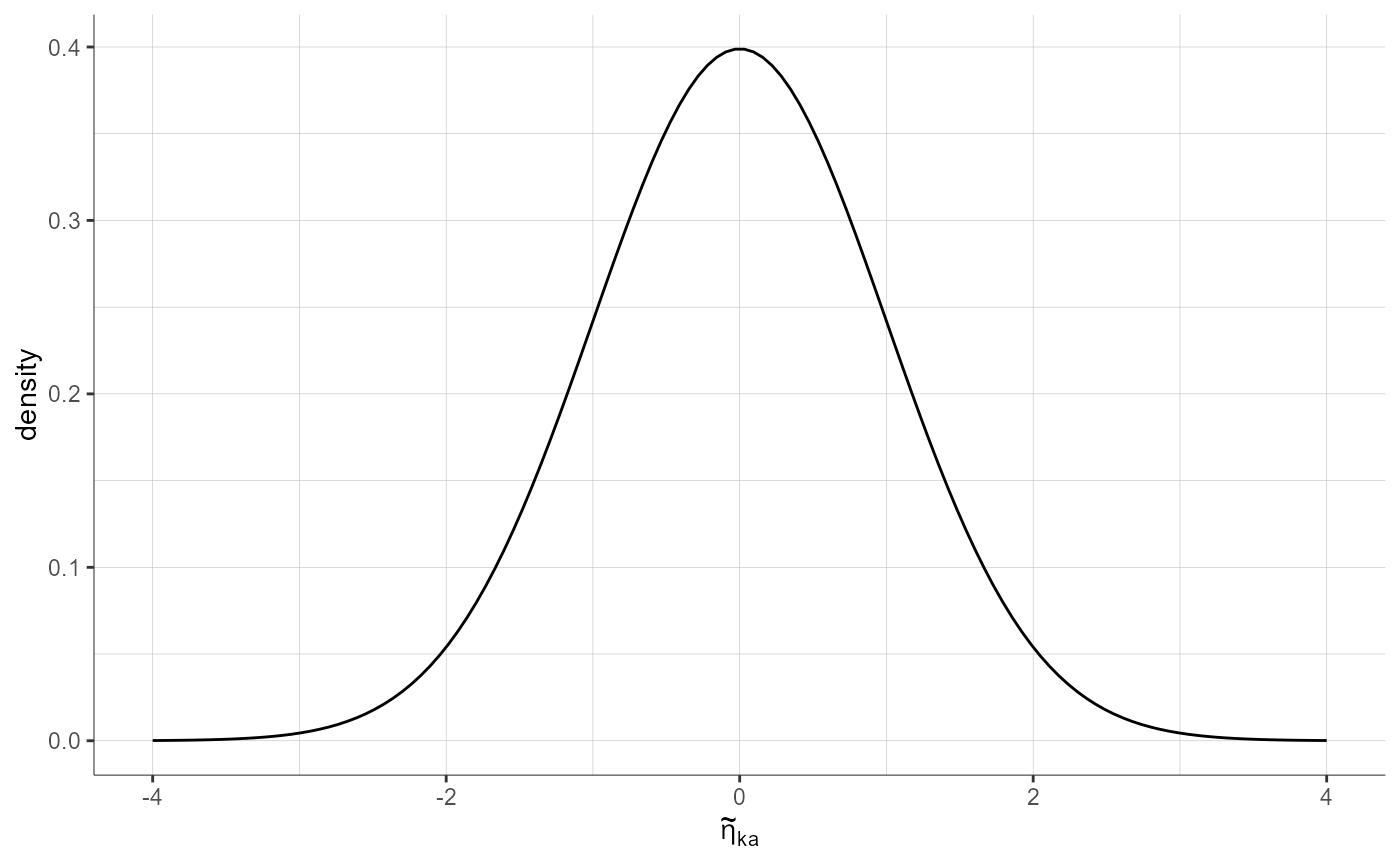
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "pdf",
settings = list(theoretical = F))
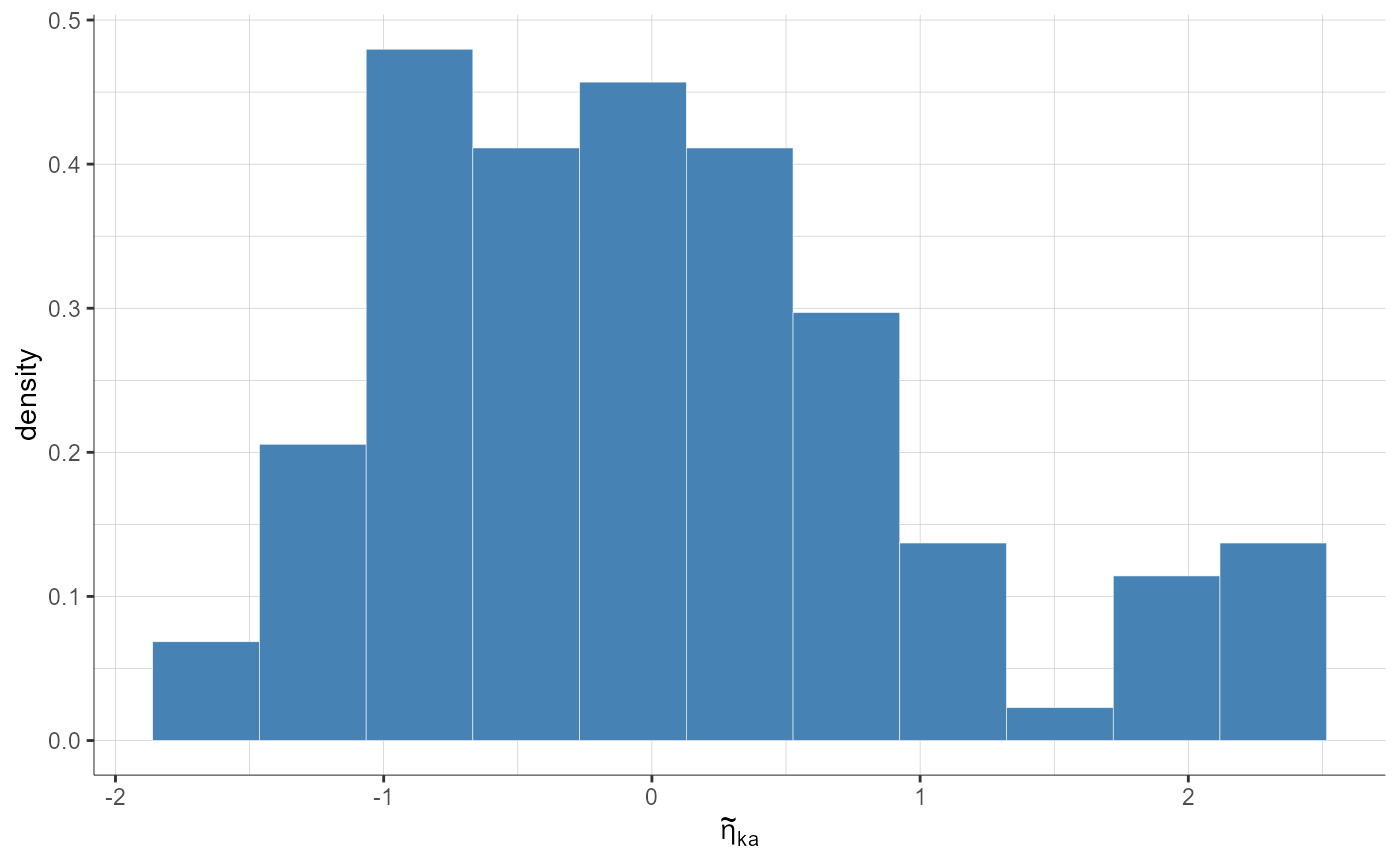
# Random effect distribution as cdf
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "cdf")
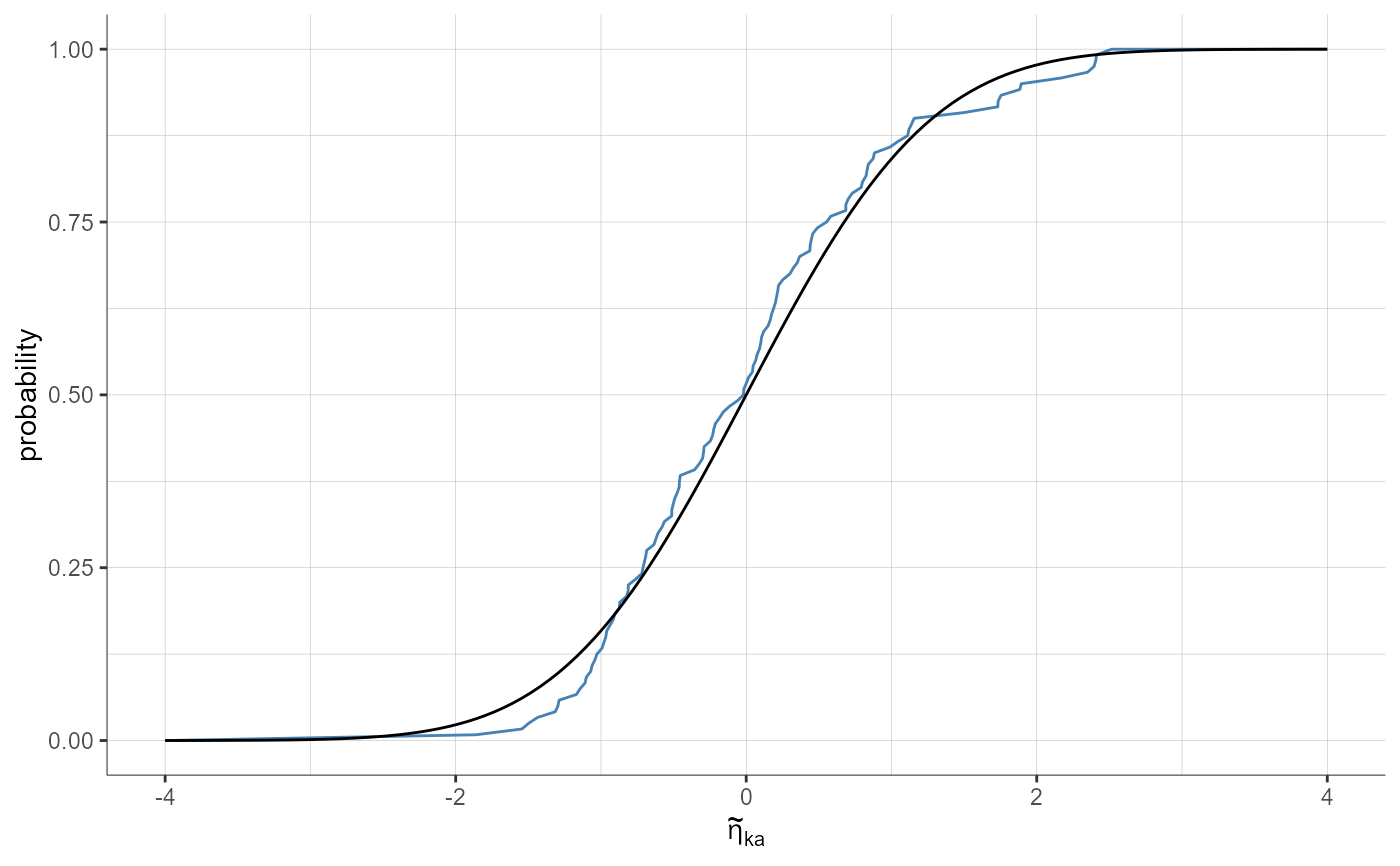
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "cdf",
settings = list(indivEstimate = "simulated"))
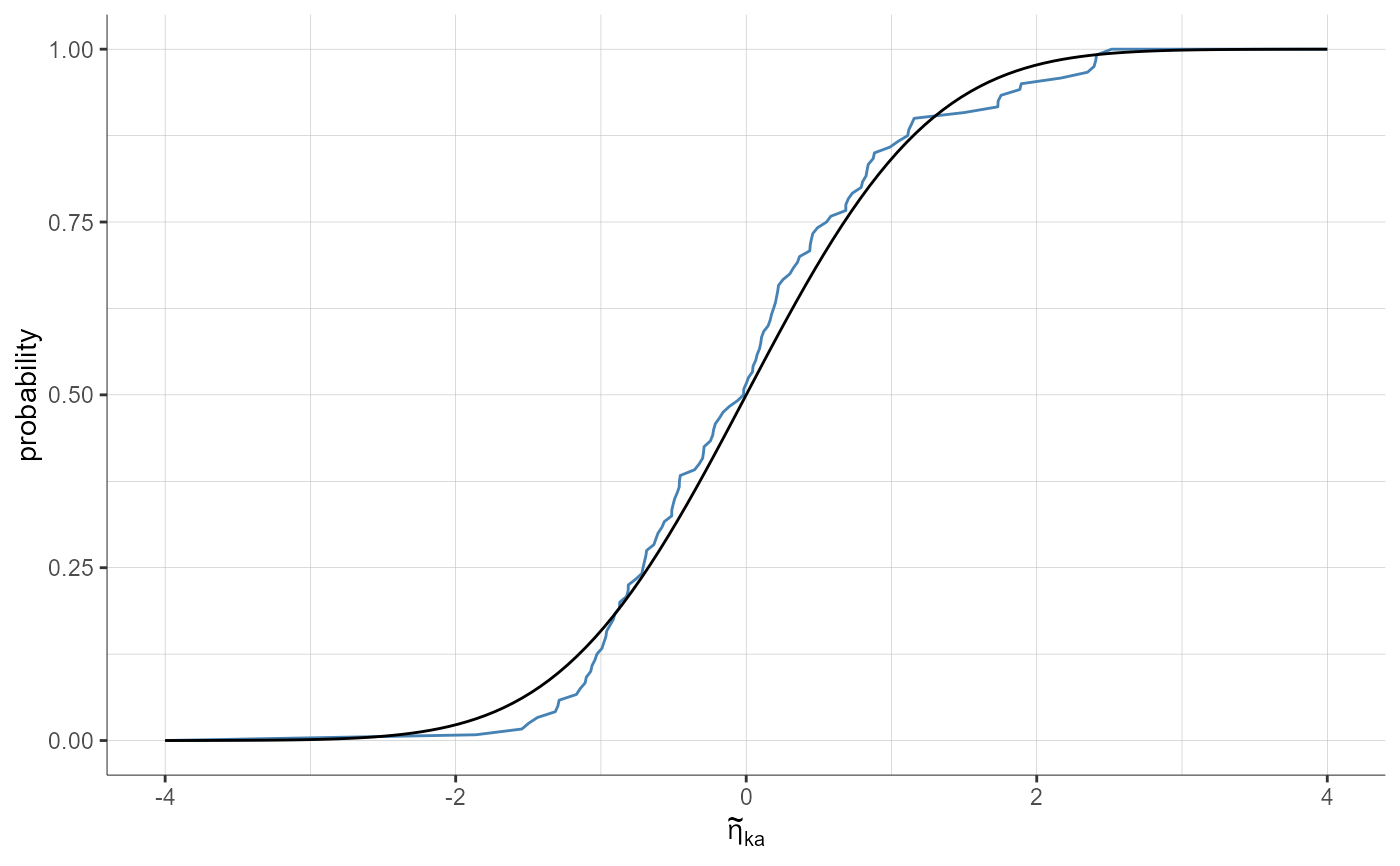
plotStandardizedRandomEffectsDistribution(parameters = "ka", plot = "cdf",
settings = list(theoretical = F))
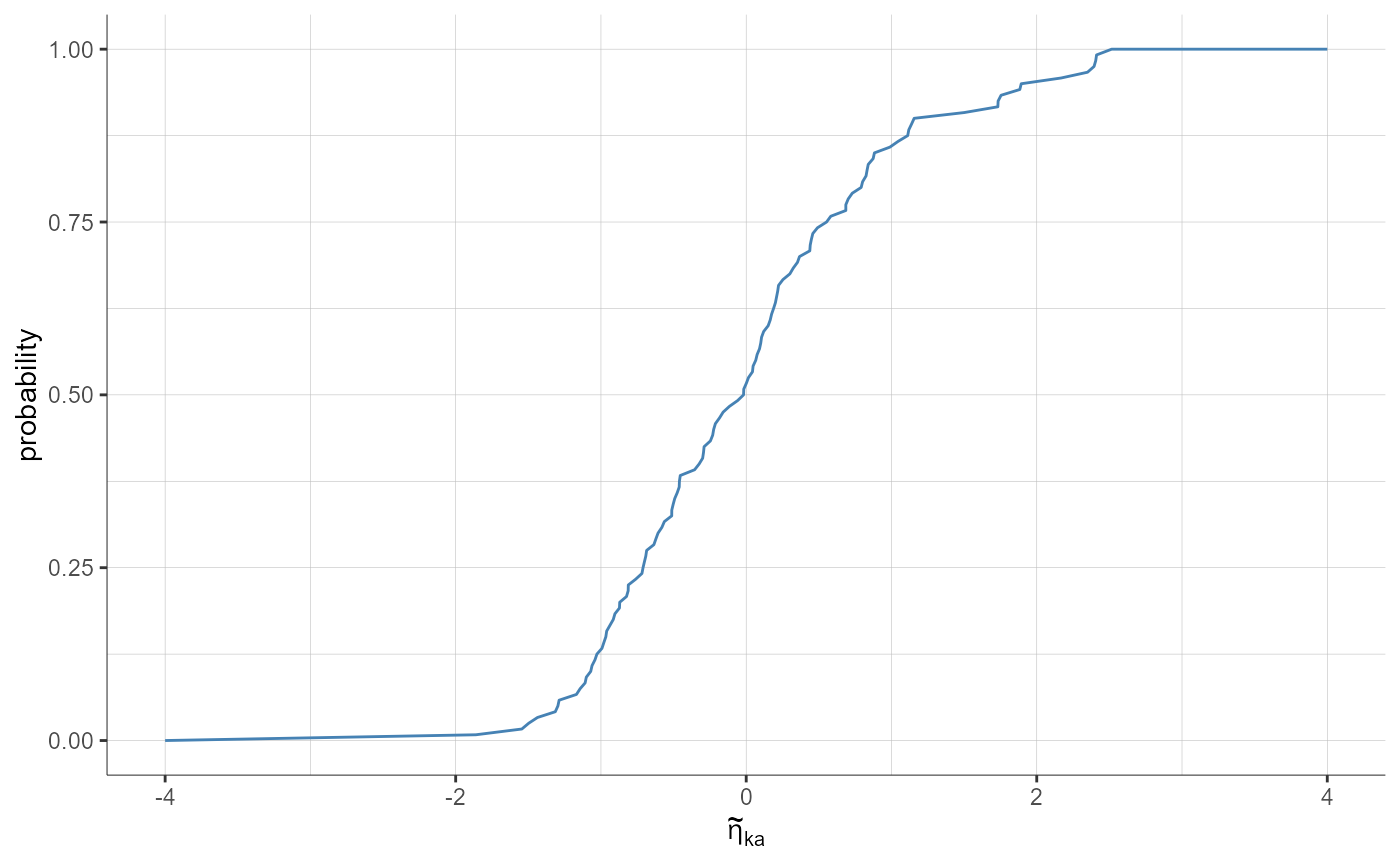
# stratification
plotStandardizedRandomEffectsDistribution(stratify = list(filter = list("WEIGHT", 1),
groups = list(name = "WEIGHT", definition = 75)))
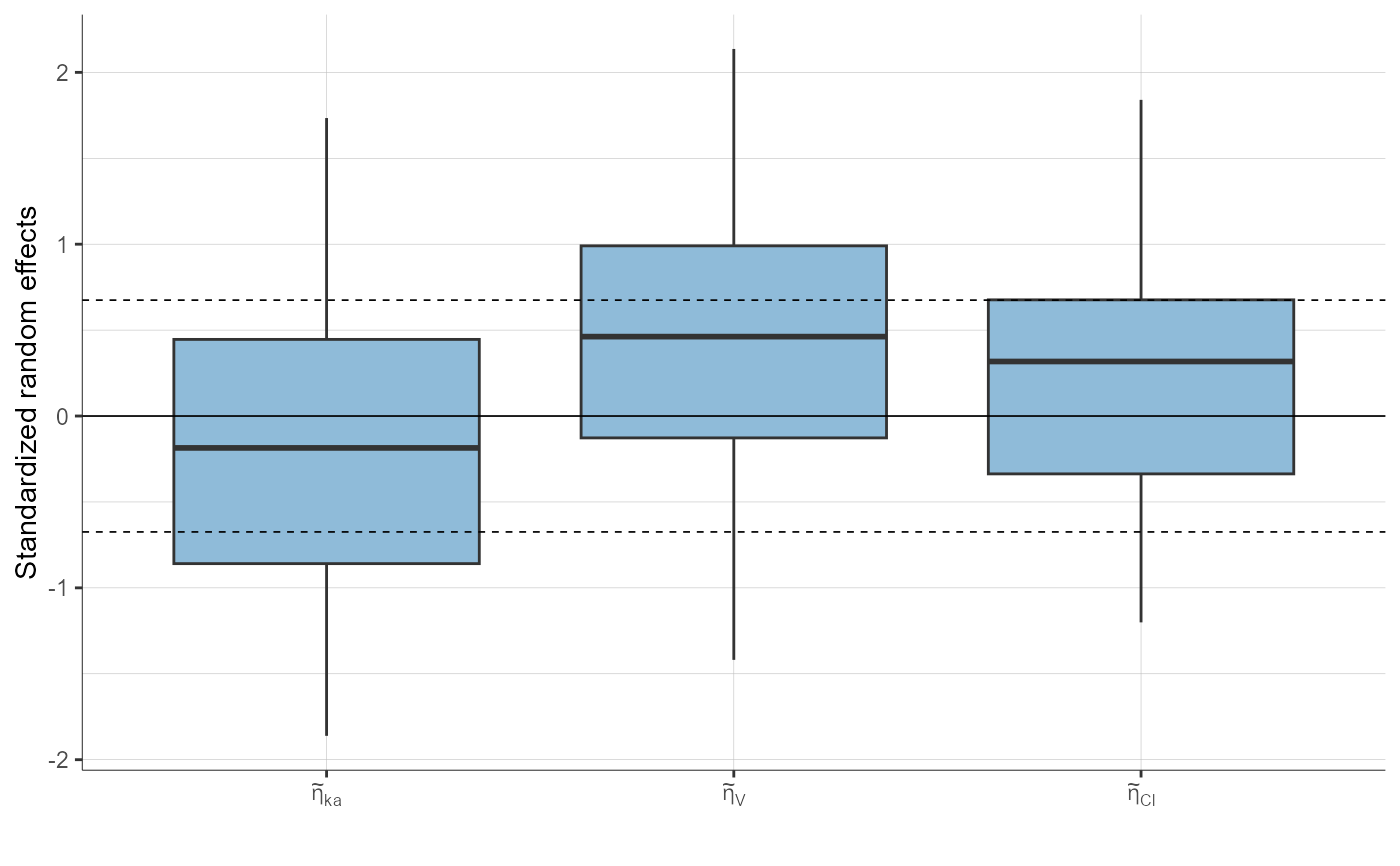
plotStandardizedRandomEffectsDistribution(parameters = "Cl",
stratify = list(split = "SEX"))
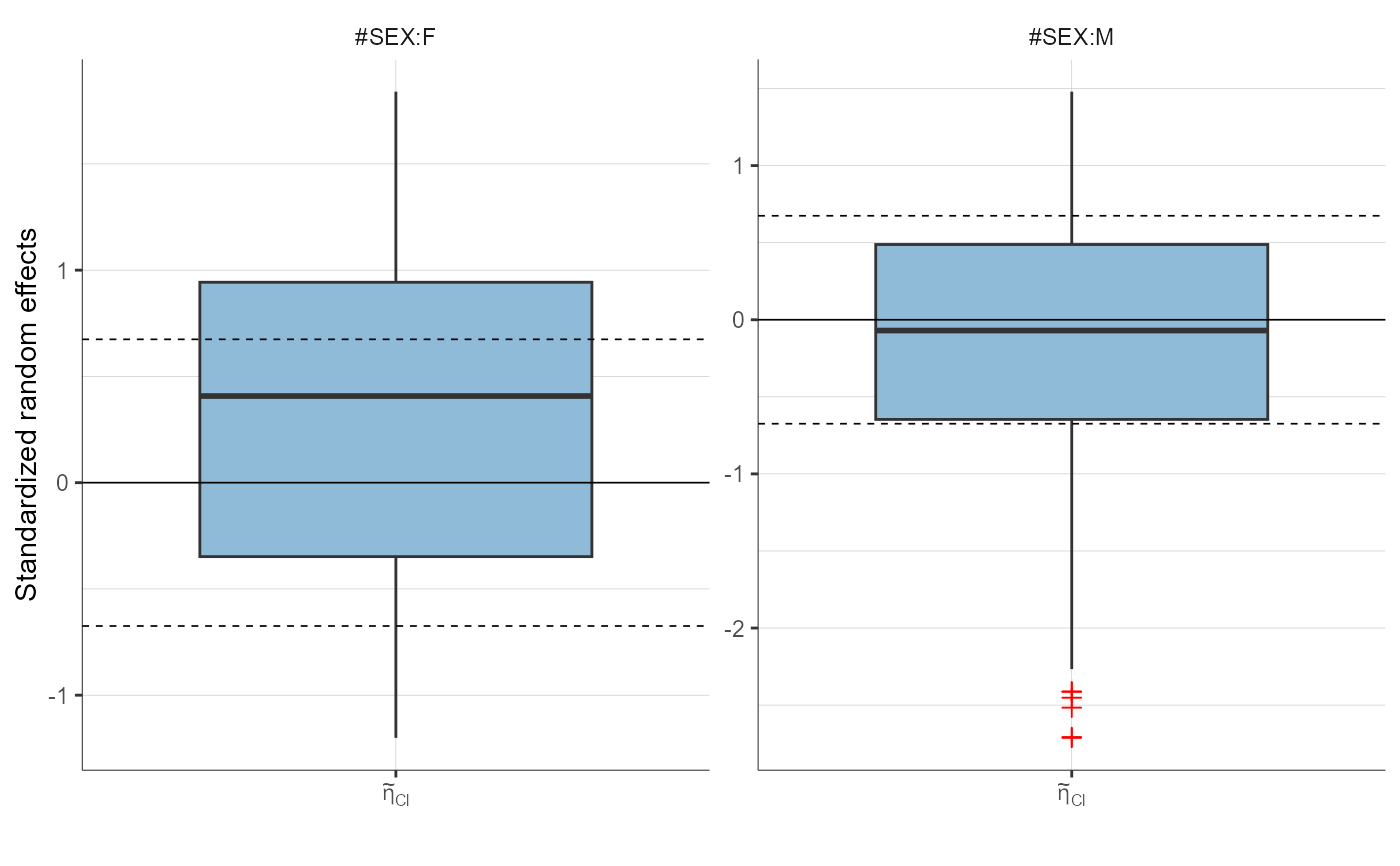
plotStandardizedRandomEffectsDistribution(parameters = c("ka", "Cl"))

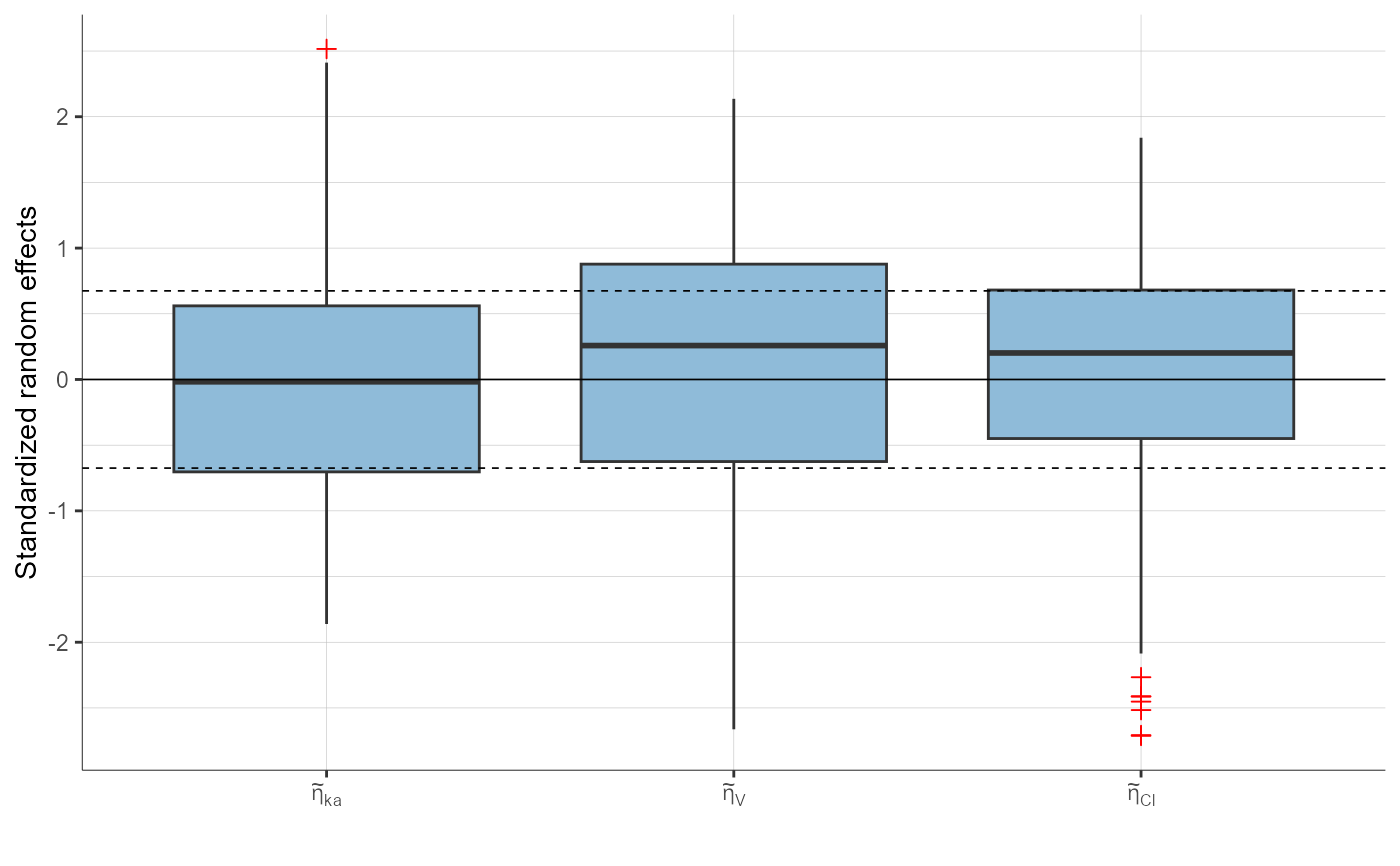
plotStandardizedRandomEffectsDistribution(plot = "boxplot")
plotStandardizedRandomEffectsDistribution(plot = "pdf")
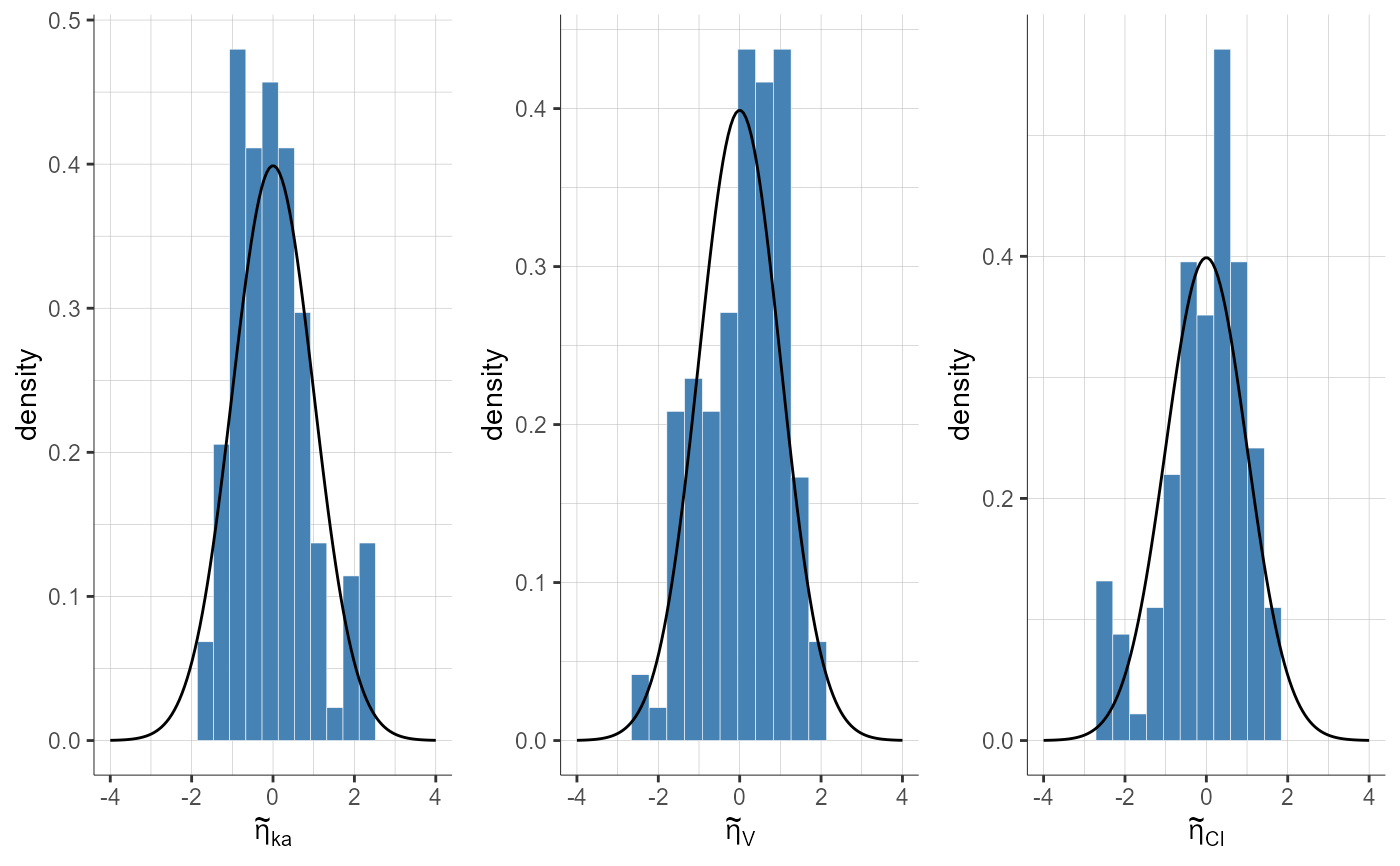
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
plotStandardizedRandomEffectsDistribution(plot = "cdf")
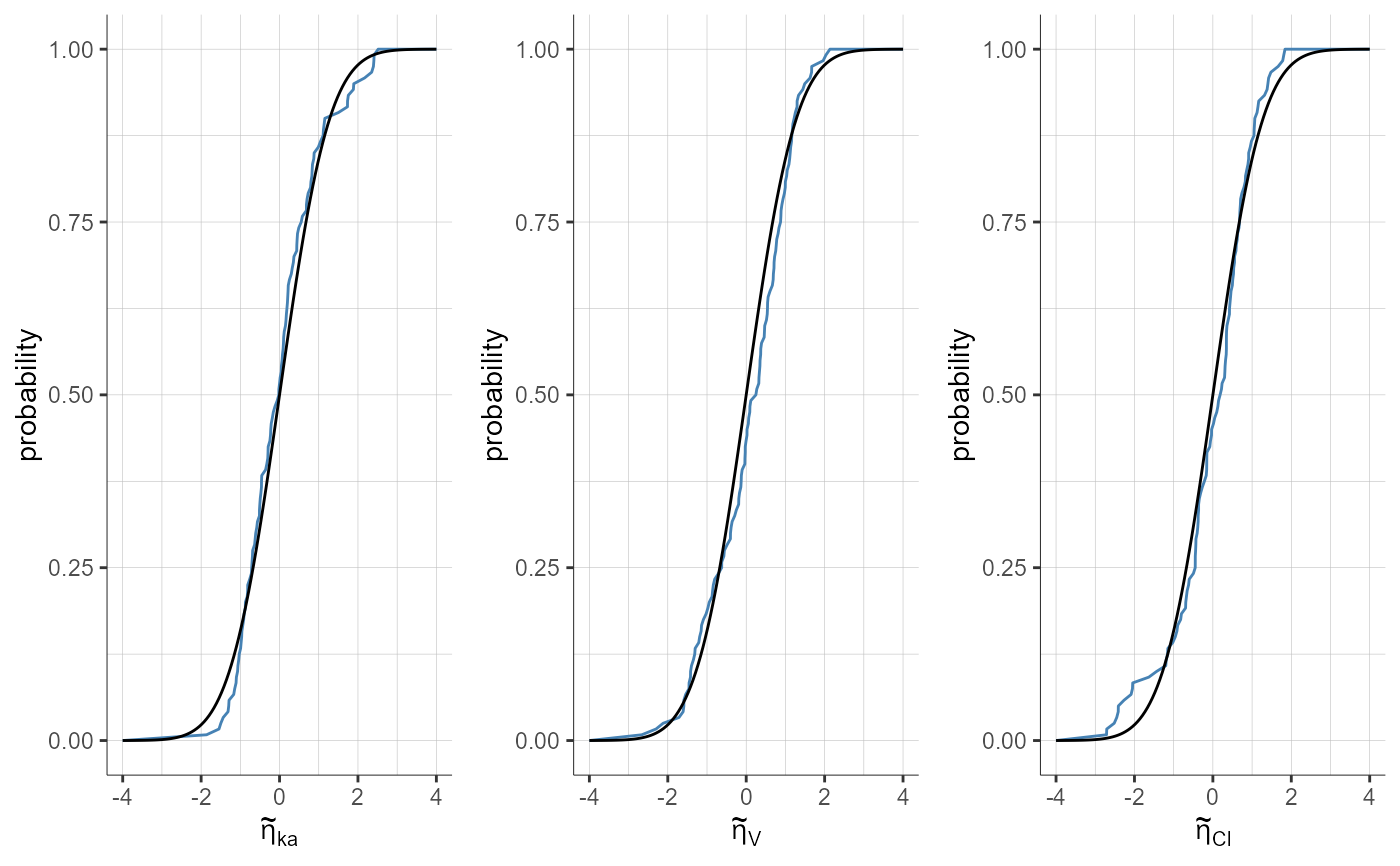
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
plotStandardizedRandomEffectsDistribution(plot = "pdf", settings = list(theoretical = F))
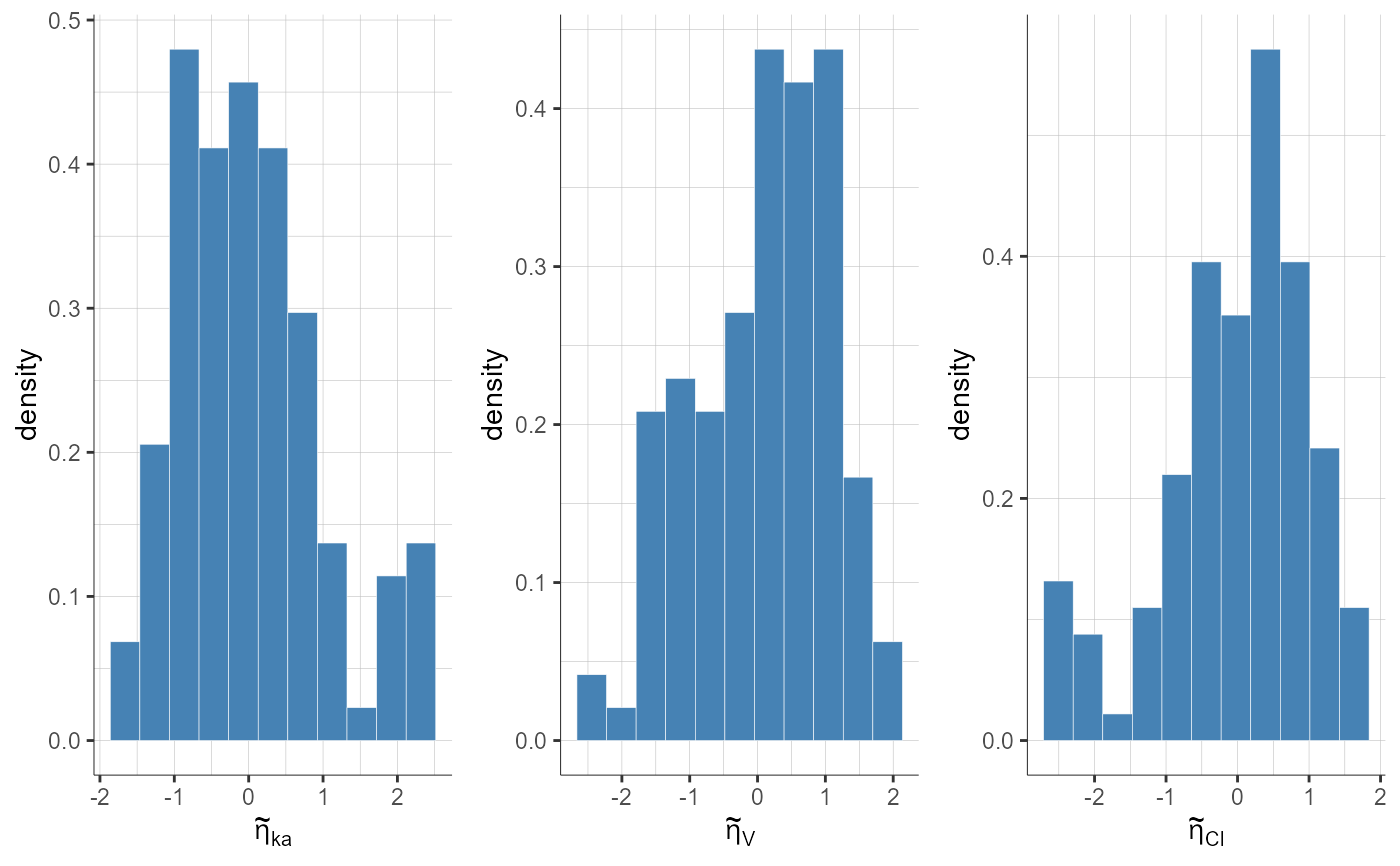
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
