plotPredictionDistribution
[Monolix] Plot distribution of the predictions
Plot the prediction distribution.
Usage
plotPredictionDistribution(
obsName = NULL,
settings = list(),
preferences = list(),
stratify = list()
)Arguments
- obsName
(character) Name of the observation (in dataset header). By default the first observation is considered.
- settings
a list of optional settings
perc(logical) - If TRUE display 9 Bands for each percentile (default TRUE).median(logical) - If TRUE display Median (default TRUE).nbBands(integer) - Number of bands to display (default 9).higherPercentile(integer) - Percentile level (default 90).obs(logical) - If TRUE display observations as dots (default FALSE).cens(logical) - If TRUE display censored observations as dots (default FALSE).binLimits(logical) If TRUE display limits of bins (default FALSE). For discrete data only.binsSettingsa list of settings for time axis binning for observation statistics computation (discrete data only):criteria(character) - Bining criteria, one of 'equalwidth', 'equalsize', or 'leastsquare' methods. (default leastsquare).is.fixedNbBins(logical) - If TRUE define a fixed number of bins, else define a range for automatic selection (default FALSE).nbBins(integer) - Define a fixed number of bins (default 10).binRange(vector(integer, integer)) - Define a range for the number of bins (default c(5, 100)).nbBinData(vector(integer, integer)) - Define a range for the number of data points per bin (default c(10, 200) for Monolix and c(3, 200) for PKanalix).
legend(logical) add (TRUE) / remove (FALSE) plot legend (default FALSE).grid(logical) add (TRUE) / remove (FALSE) plot grid (default TRUE).xlog(logical) add (TRUE) / remove (FALSE) log scaling on x axis (default FALSE).ylog(logical) add (TRUE) / remove (FALSE) log scaling on y axis (default FALSE).xlab(character) label on x axis (default "Time").ylab(character) label on y axis (default obsName).ncol(integer) number of columns when facet = TRUE (default 4).xlim(c(double, double)) limits of the x axis.ylim(c(double, double)) limits of the y axis.fontsize(integer) Plot text font size.scales(character) Should scales be fixed ("fixed"), free ("free", the default), or free in one dimension ("free_x", "free_y") (default "free").
- preferences
(optional) preferences for plot display, run getPlotPreferences("plotPredictionDistribution") to check available displays.
- stratify
List with the stratification arguments:
groups- Definition of stratification groups. By default, stratification groups are already defined as one group for each category for categorical covariates, and two groups of equal number of individuals for continuous covariates. To redefine groups, for each covariate to redefine, specify a list with:name character covariate name (e.g "AGE")definition (vector(continuous) || list>(categorical)) For continuous covariates, vector of break values (e.g c(35, 65)). For categorical covariates, groups of categories as a list of vectors(e.glist(c("study101"), c("study201","study202")))split(vector) - Vector of covariates used to split (i.e facet) the plot (by default no split is applied). For instancec("FORM","AGE").filter(list< list> >) - List of pairs containing a covariate name and the vector of indexes or categories (for categorical covariates) of the groups to keep (by default no filtering is applied). For instance,list("AGE",c(1,3))to keep the individuals belonging to the first and third age group, according to the definition ingroups. For instance,list("FORM","ref")using the category name for categorical covariates.color(vector) - Vector of covariates used for coloring (by default no coloring is applied). For instancec("FORM","AGE").colors- Vector of colors to use whencolorargument is used. Takes precedence over colors defined inpreferences. For instancec("#ebecf0","#cdced1","#97989c").
Value
a ggplot2 object
Details
Note that computation settings are not available for this connector in 2021 version: Number of bands is set to 9 and Level is set to 90
Note that stratification options are not available for this connector in 2021 version:
See also
Examples
initializeLixoftConnectors(software = "monolix")
# continuous data
project <- file.path(getDemoPath(), "1.creating_and_using_models",
"1.1.libraries_of_models", "theophylline_project.mlxtran")
loadProject(project)
runPopulationParameterEstimation()
plotPredictionDistribution()
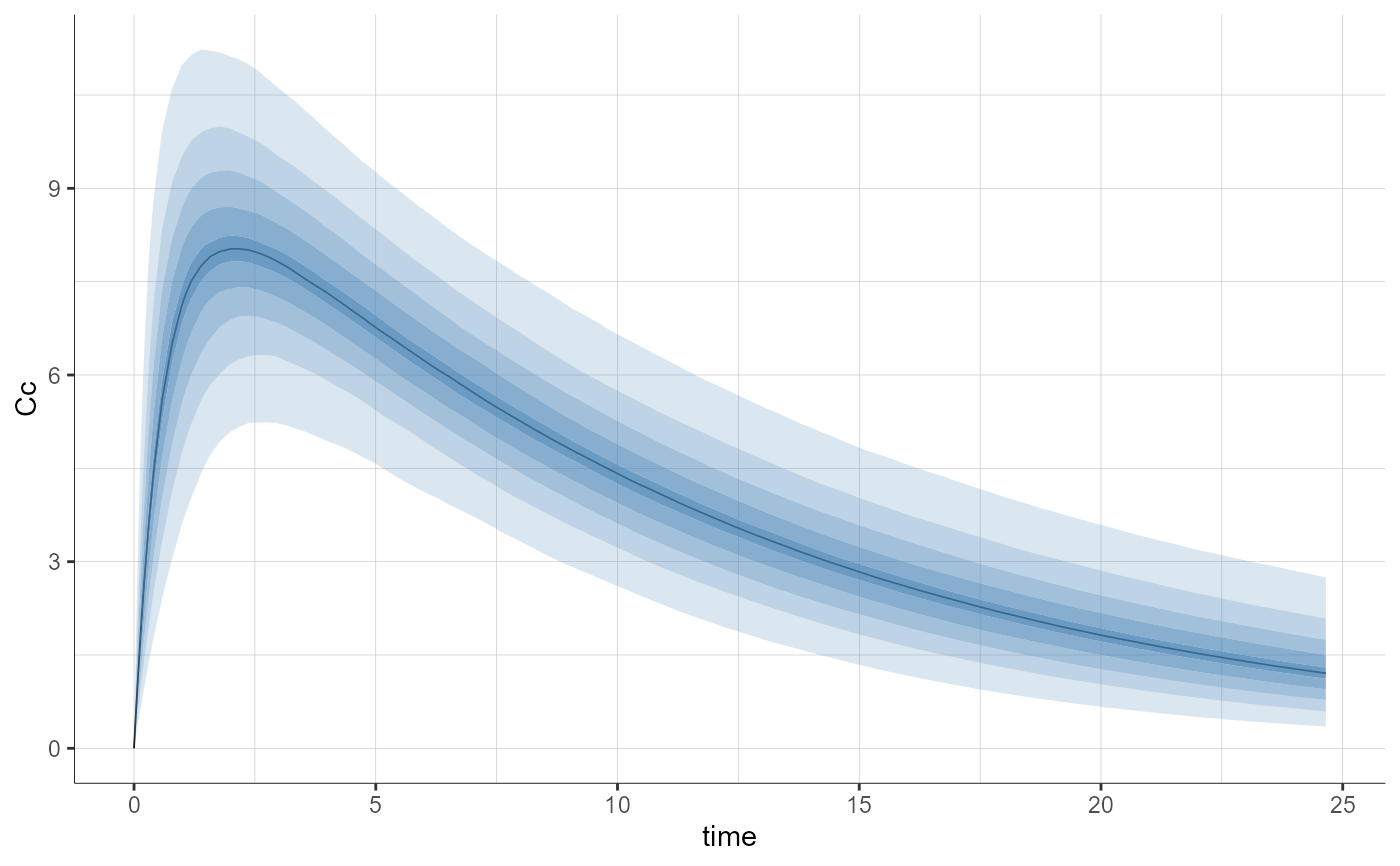
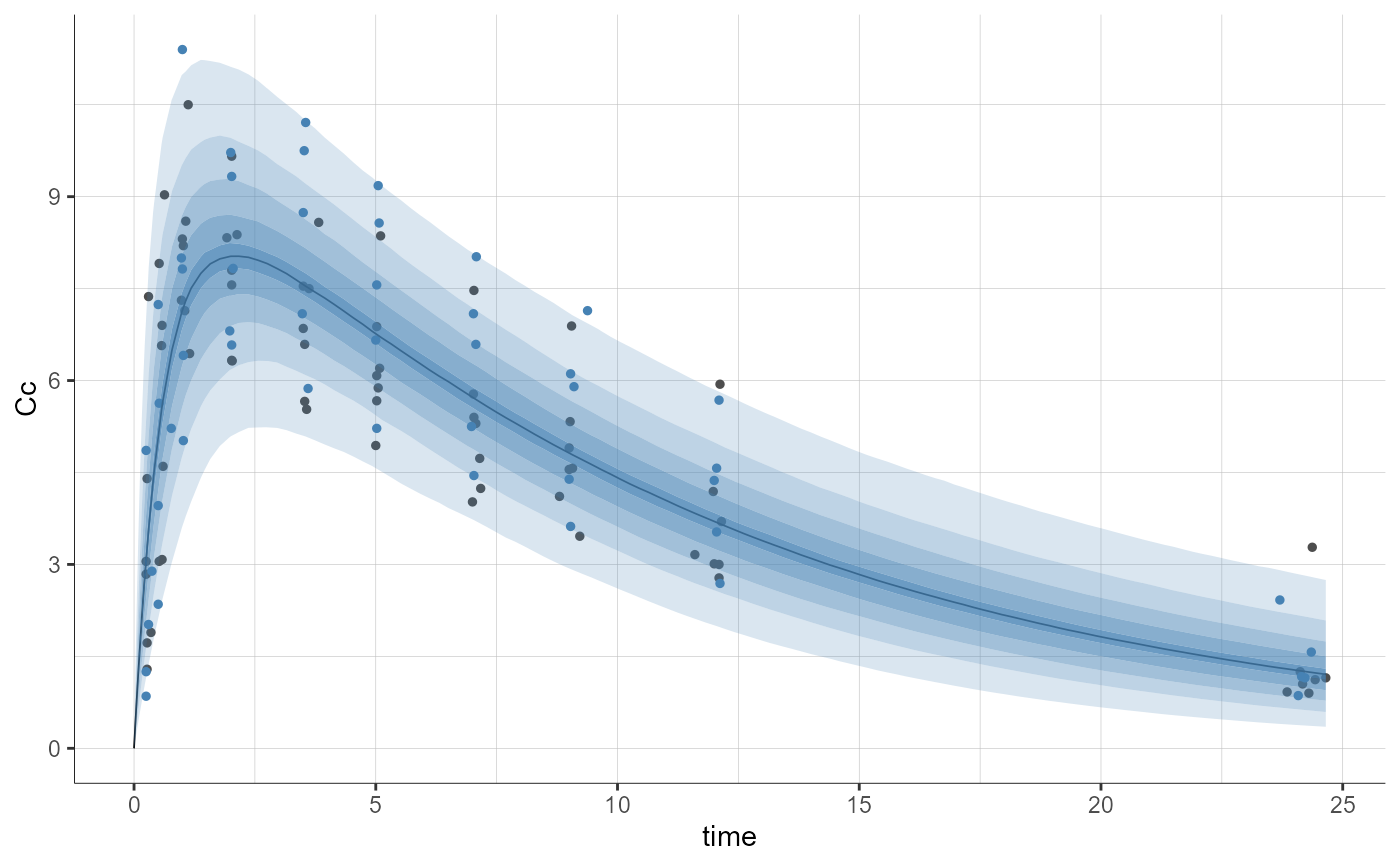
plotPredictionDistribution(stratify = list(color = "SEX"), settings = list(obs = TRUE))
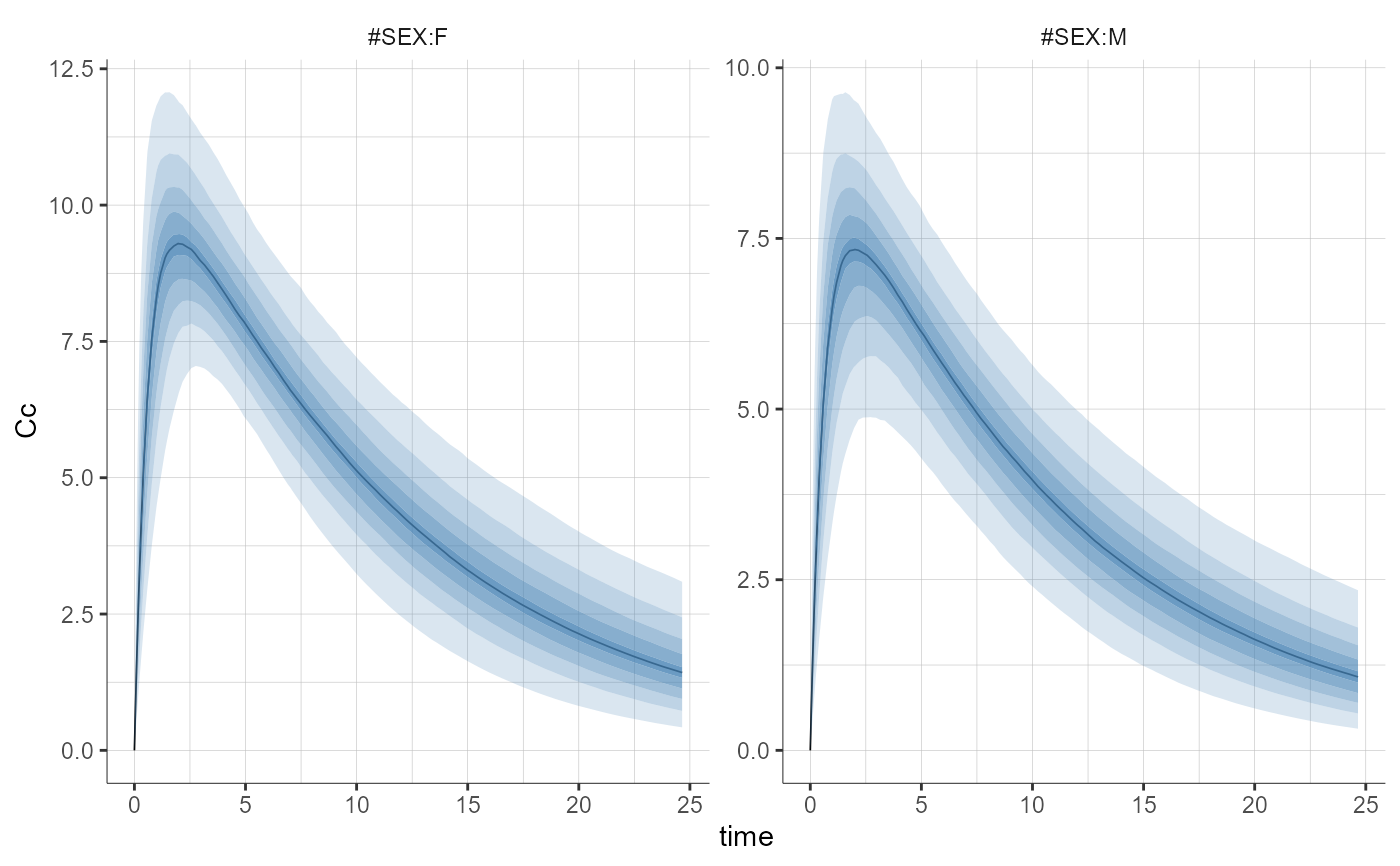
plotPredictionDistribution(stratify = list(split = "SEX"))
$body