plotCovariates
[Monolix - PKanalix] Generate Covariate plots
Plot the covariates.
Usage
plotCovariates(
covariatesRows = NULL,
covariatesColumns = NULL,
settings = list(),
preferences = list(),
stratify = list()
)Arguments
- covariatesRows
vector with the name of covariates to display on rows (by default the first 4 covariates are displayed).
- covariatesColumns
vector with the name of covariates to display on columns (by default the first 4 covariates are displayed).
- settings
List with the following settings
regressionLine(logical) If TRUE, Add regression line in scatterplots (default TRUE).spline(logical) If TRUE, Add xpline in scatterplots (default FALSE).histogramColors(vector) List of colors to use in histograms plots.histogramPosition(character) Type of histogram: "stacked", "grouped" or "default" (histograms with categorical covariates in xaxis the plot is grouped else it is stacked), (Default is "default")legend(logical) add (TRUE) / remove (FALSE) plot legend (default FALSE).grid(logical) add (TRUE) / remove (FALSE) plot grid (default TRUE).ncol(integer) number of columns when facet = TRUE (default 4).bins(integer) number of bins for the histogram (default 10)fontsize(integer) Plot text font size.
- preferences
(optional) preferences for plot display, run getPlotPreferences("plotCovariates") to check available displays.
- stratify
List with the stratification arguments:
groups- Definition of stratification groups. By default, stratification groups are already defined as one group for each category for categorical covariates, and two groups of equal number of individuals for continuous covariates. To redefine groups, for each covariate to redefine, specify a list with:name character covariate name (e.g "AGE")definition (vector(continuous) || list>(categorical)) For continuous covariates, vector of break values (e.g c(35, 65)). For categorical covariates, groups of categories as a list of vectors(e.glist(c("study101"), c("study201","study202")))split(vector) - Vector of covariates used to split (i.e facet) the plot (by default no split is applied). For instancec("FORM","AGE").filter(list< list> >) - List of pairs containing a covariate name and the vector of indexes or categories (for categorical covariates) of the groups to keep (by default no filtering is applied). For instance,list("AGE",c(1,3))to keep the individuals belonging to the first and third age group, according to the definition ingroups. For instance,list("FORM","ref")using the category name for categorical covariates.color(vector) - Vector of covariates used for coloring (by default no coloring is applied). For instancec("FORM","AGE").colors- Vector of colors to use whencolorargument is used. Takes precedence over colors defined inpreferences. For instancec("#ebecf0","#cdced1","#97989c").individualSelection- Ids to display (by default the 12 first ids are displayed) defined as:indices(vector) - Indices of the individuals to display (by default, the 12 first individuals are selected). If occasions are present, all occasions of the selected individuals will be displayed. Takes precedence overids. For instancec(5,6,10,11).isRange(logical) - If TRUE, all individuals whose index is inside [min(indices), max[indices]] are selected (FALSE by default). Forced to FALSE ifidsis defined.ids(vector) - Names of the individuals to display. If occasions are present, all occasions of the selected individuals will be displayed. For instancec("101-01","101-02","101-03"). If ids are integers, can also bec(1,3,6). Ignored ifindicesis defined.
Value
A ggplot object if one element in covariatesRows and covariatesColumns,
A TableGrob object if multiple plots (output of grid.arrange)
Details
Generate scatterplots between two continuous covariates or bar plot between categorical covariates.
See also
Examples
initializeLixoftConnectors(software = "pkanalix")
project <- file.path(getDemoPath(), "2.case_studies/project_Theo_extravasc_SD.pkx")
loadProject(project)
# covariate distribution when only one covariate is specified
plotCovariates(covariatesRows = "HT", settings = list(bins = 10))
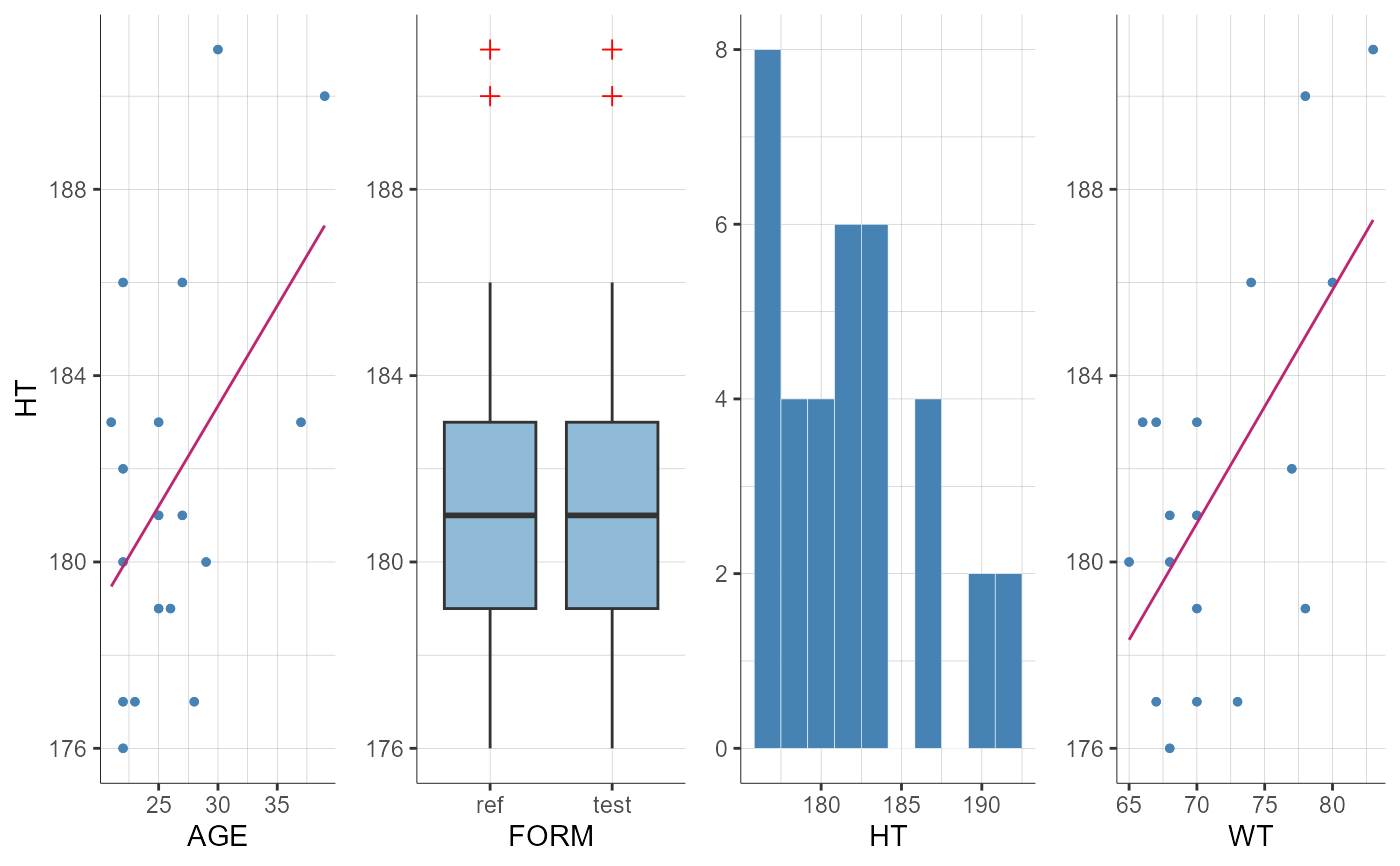
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
# scatter plot when both covariates are continuous
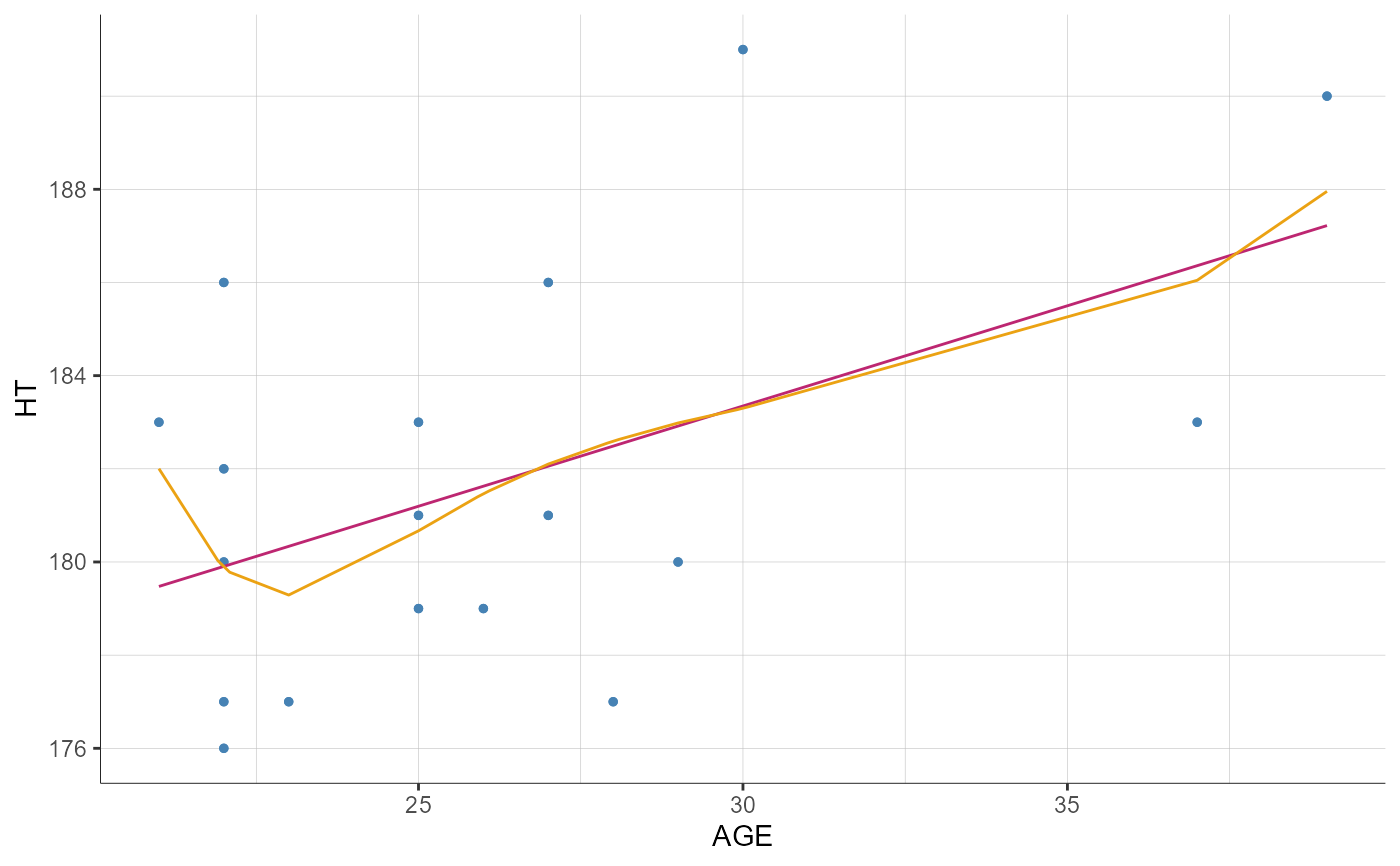
plotCovariates(covariatesRows = "HT", covariatesColumns = "AGE", settings = list(spline = TRUE))
plotCovariates(covariatesRows = "HT", covariatesColumns = c("AGE", "FORM"))
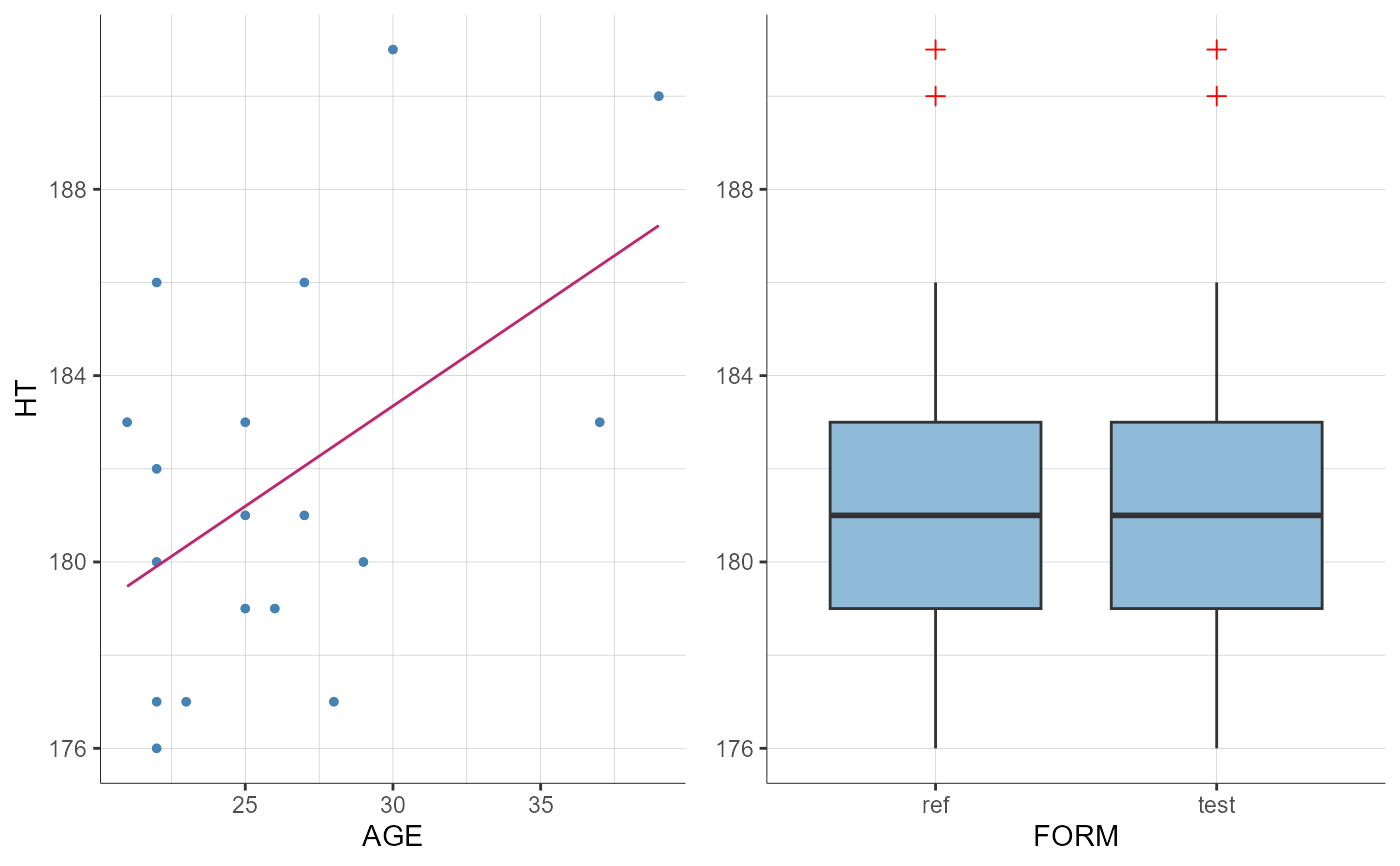
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
# box plot when one covariate is categorical and the othe one is continuous
preferences <- list(boxplot = list(fill = "#2075AE"), boxplotOutlier = list(shape = 3))
plotCovariates(covariatesRows = "FORM", covariatesColumns = "AGE", preferences = preferences)
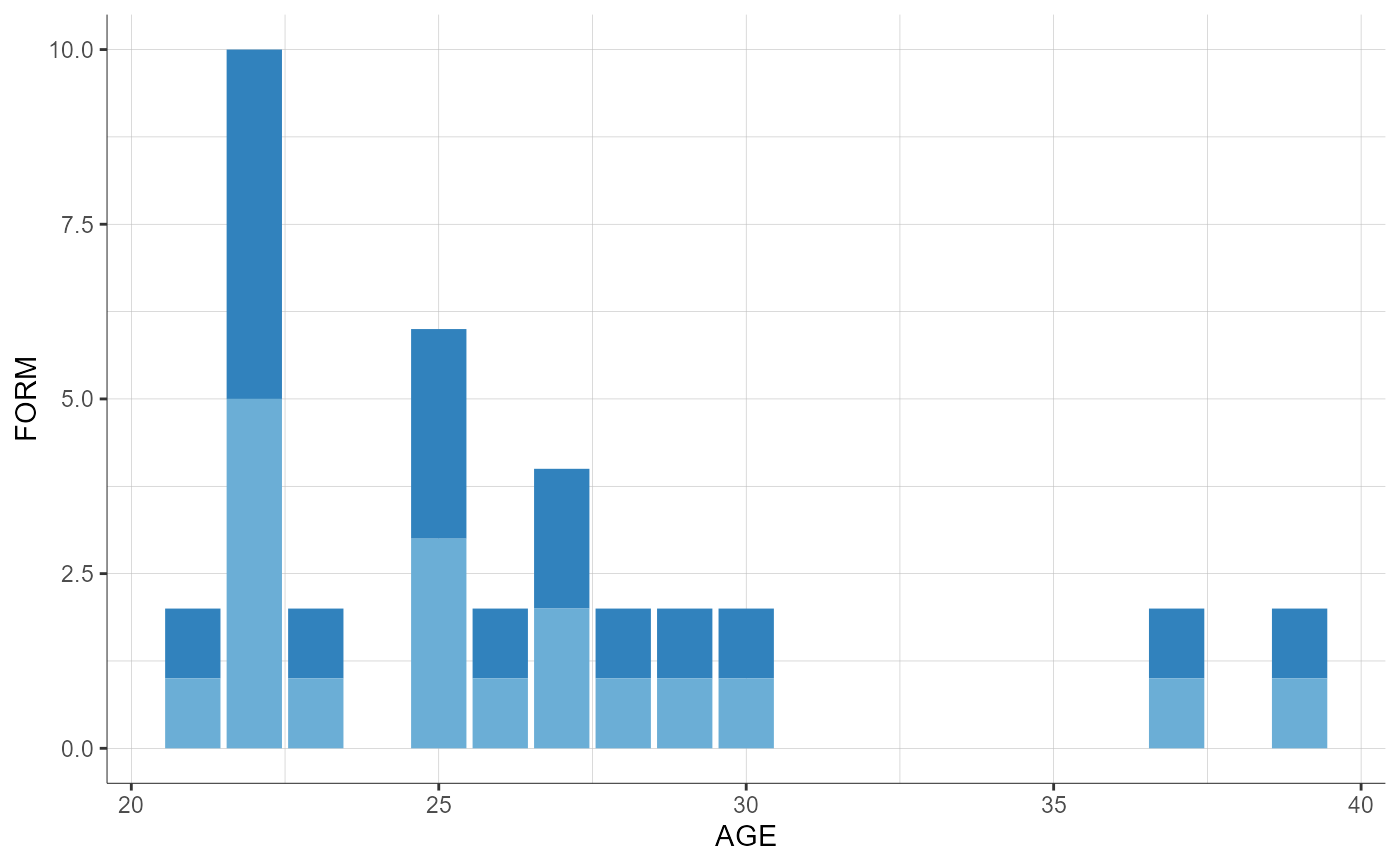
# histogram when covariate on column is categorical
plotCovariates(covariatesRows = "FORM", covariatesColumns = "SEQ",
settings = list(histogramColors = c("#5DC088", "#DBA92B")))
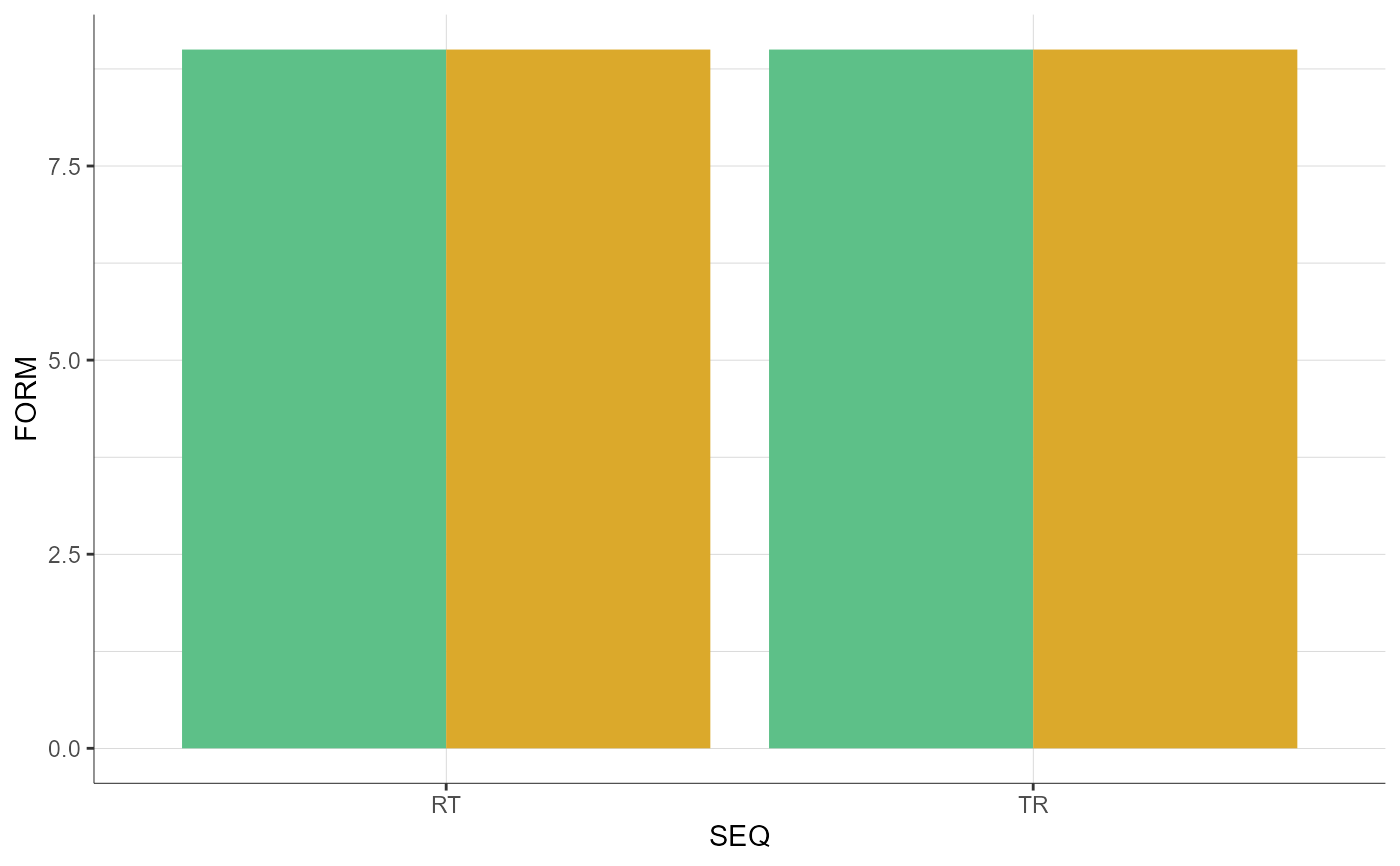
plotCovariates(covariatesRows = "AGE", covariatesColumns = "SEQ",
settings = list(histogramColors = c("#5DC088", "#DBA92B")))
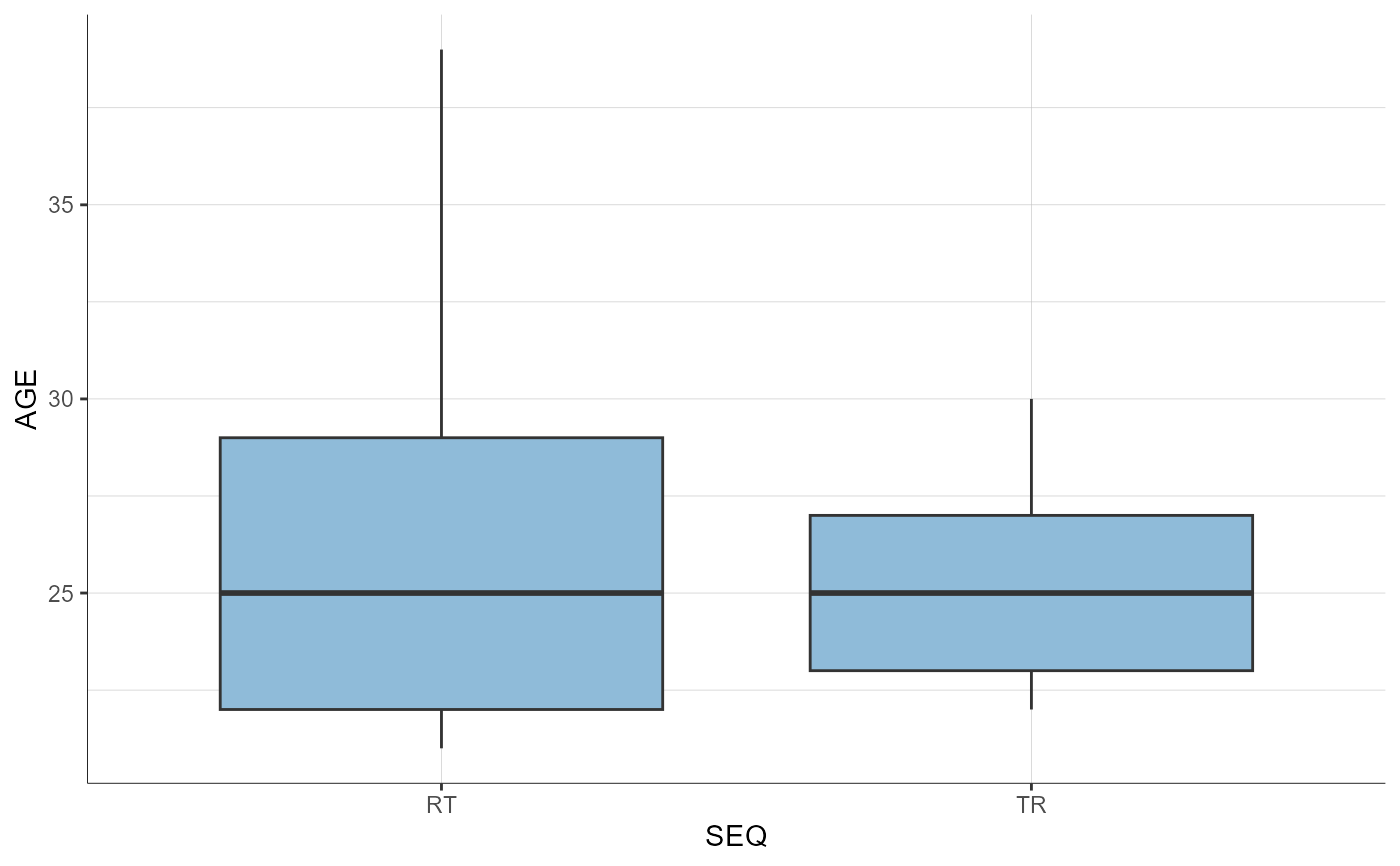
# stratification
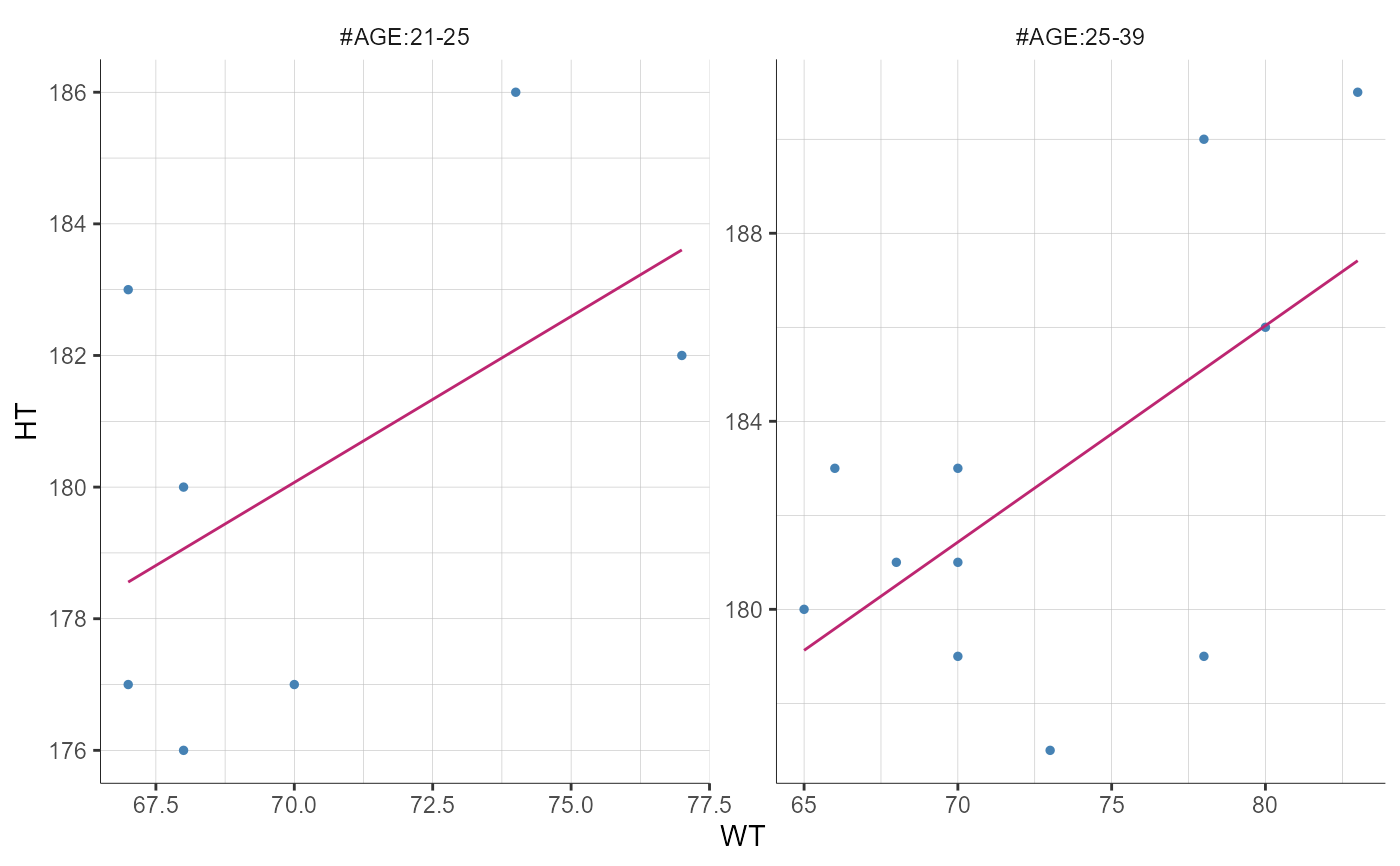
plotCovariates(covariatesRows = "HT", covariatesColumns = "WT",
stratify = list(split = "AGE",
filter = list("Period", 1),
groups = list(name = "AGE", definition = 25)))
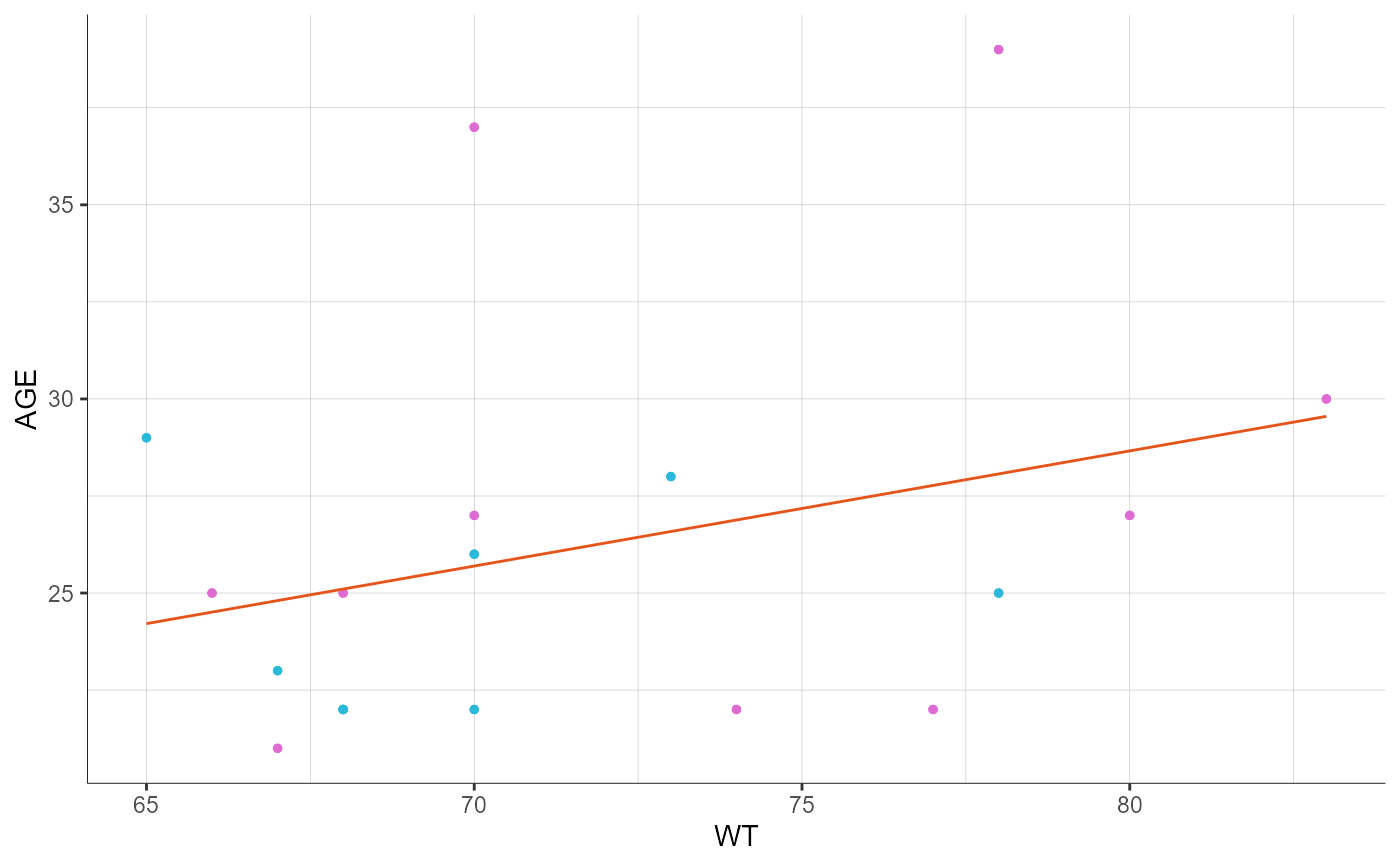
preferences <- list(regressionLine = list(color = "#E5551B"))
plotCovariates(covariatesRows = "AGE", covariatesColumns = "WT",
stratify = list(color = "HT",
groups = list(name = "HT", definition = 181),
colors = c("#2BB9DB", "#DD6BD2")),
preferences = preferences)
plotCovariates(covariatesRows = "HT", covariatesColumns = "WT",
stratify = list(split = c("AGE", "SEQ"),
groups = list(name = "AGE", definition = 25)))
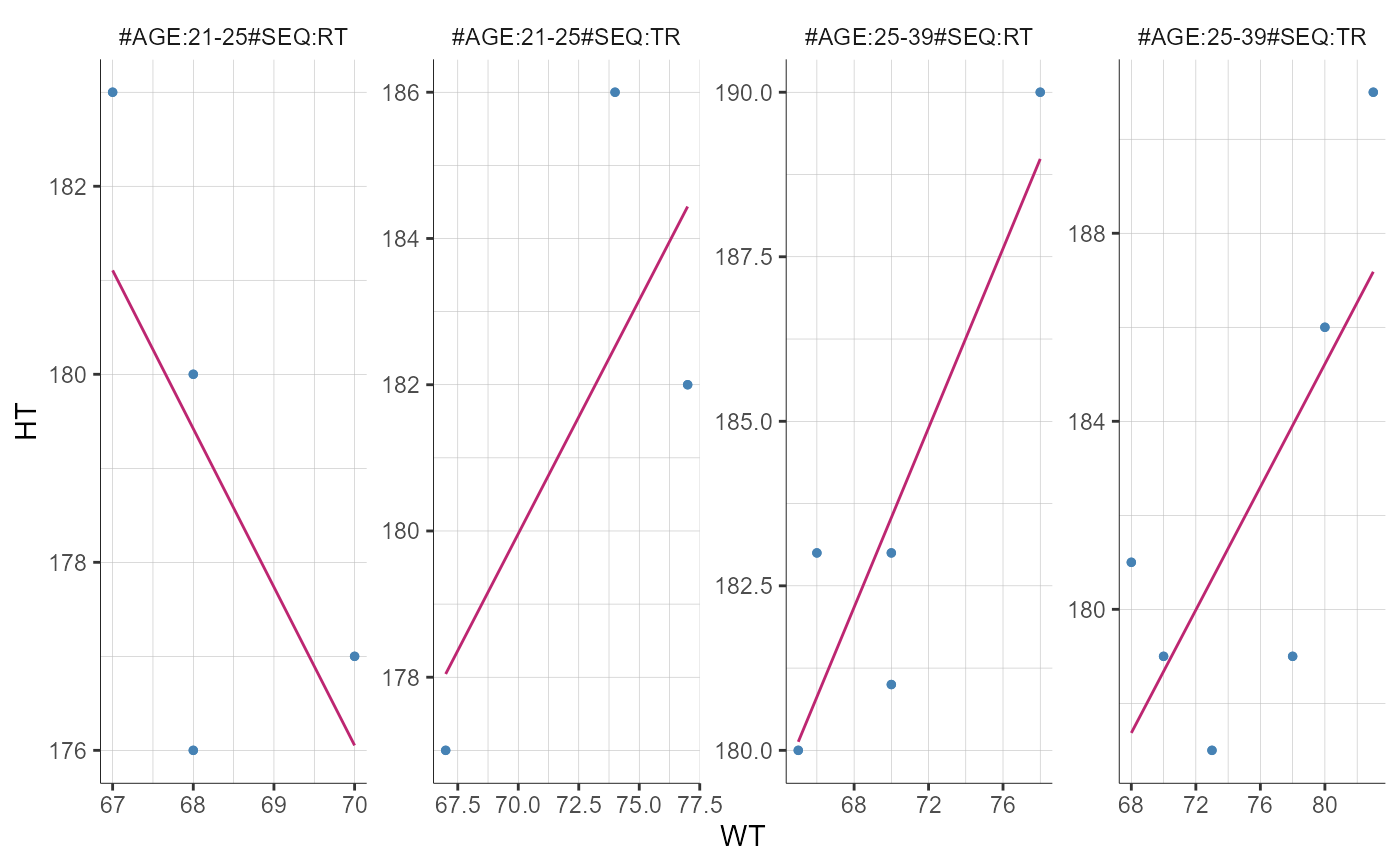
# Mulitple covariates
plotCovariates()
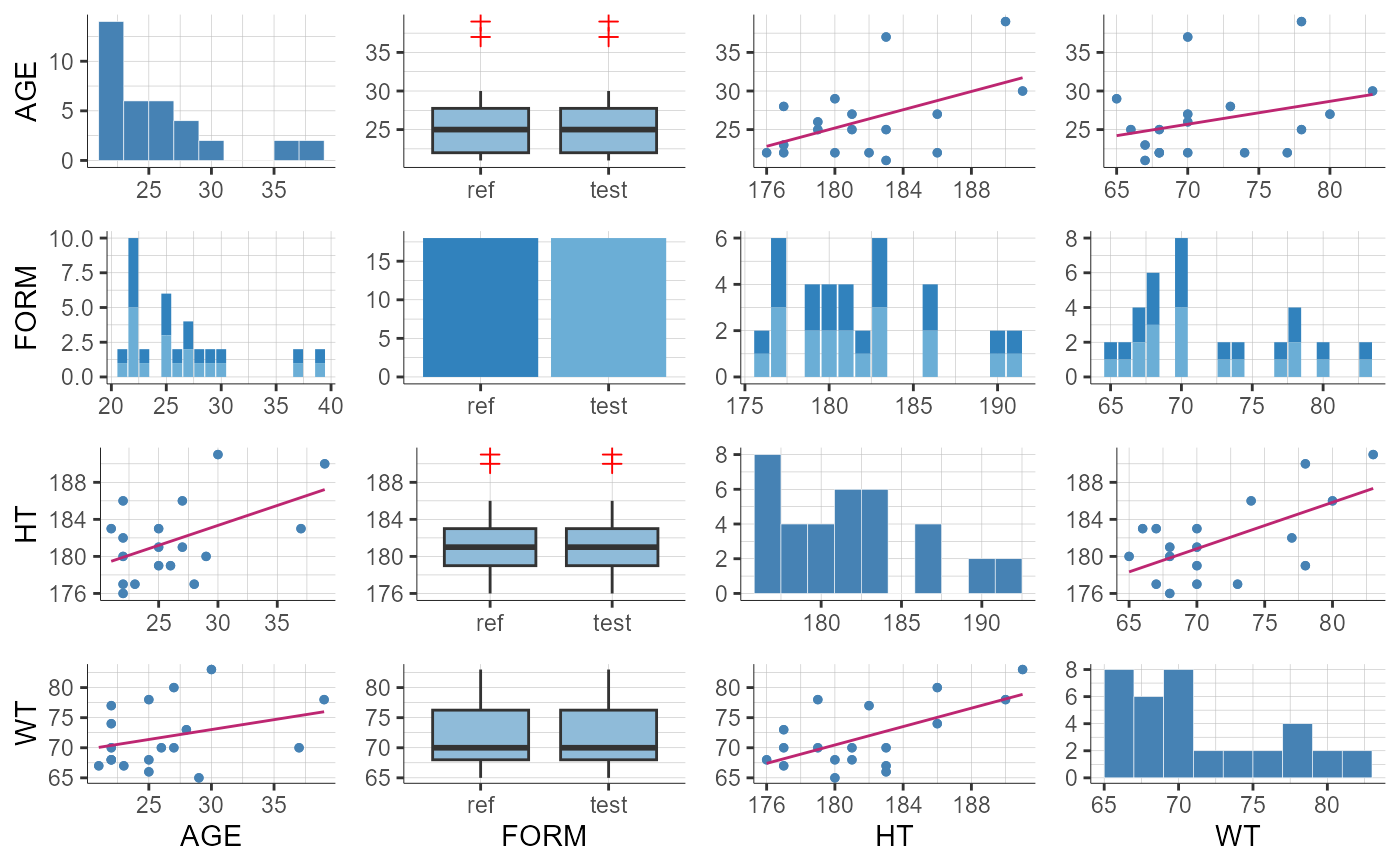
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
plotCovariates(covariatesRows = c("AGE", "SEQ", "HT"), covariatesColumns = c("AGE", "SEQ", "HT"))
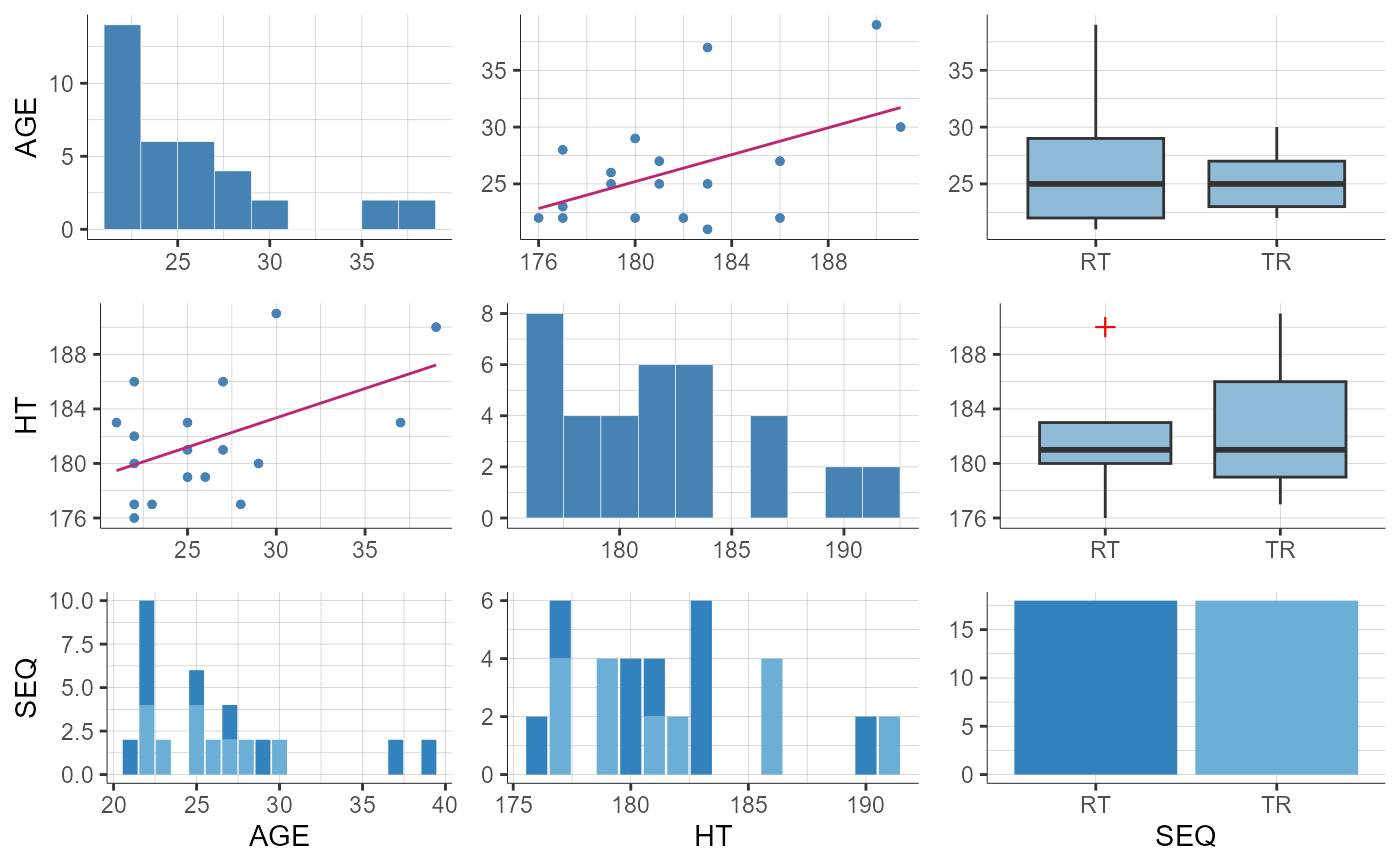
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
plotCovariates(stratify = list(filter = list("AGE", 2),
groups = list(name = "AGE", definition = c(25, 30))))
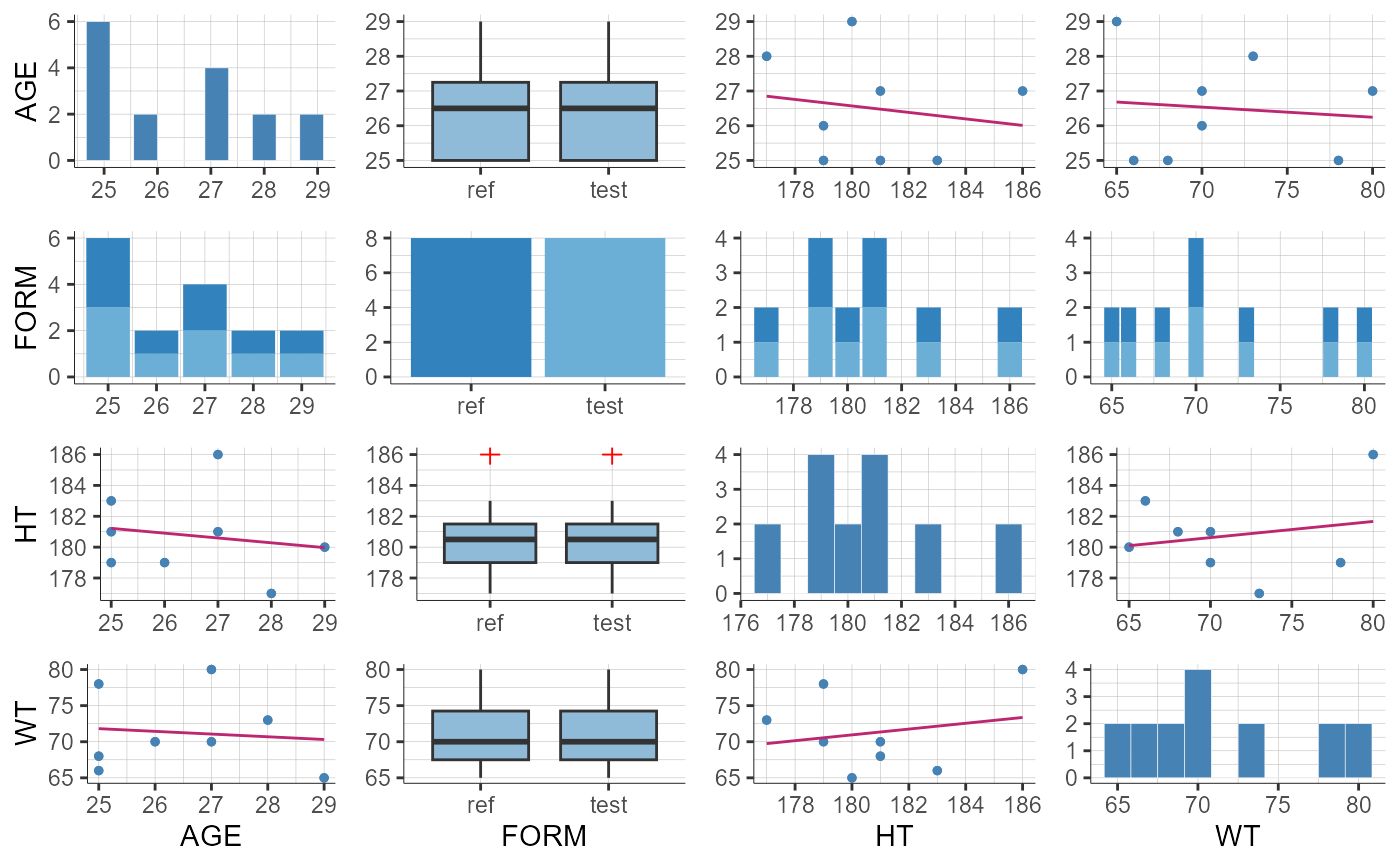
#> TableGrob (1 x 1) "arrange": 1 grobs
#> z cells name grob
#> 1 1 (1-1,1-1) arrange gtable[arrange]
