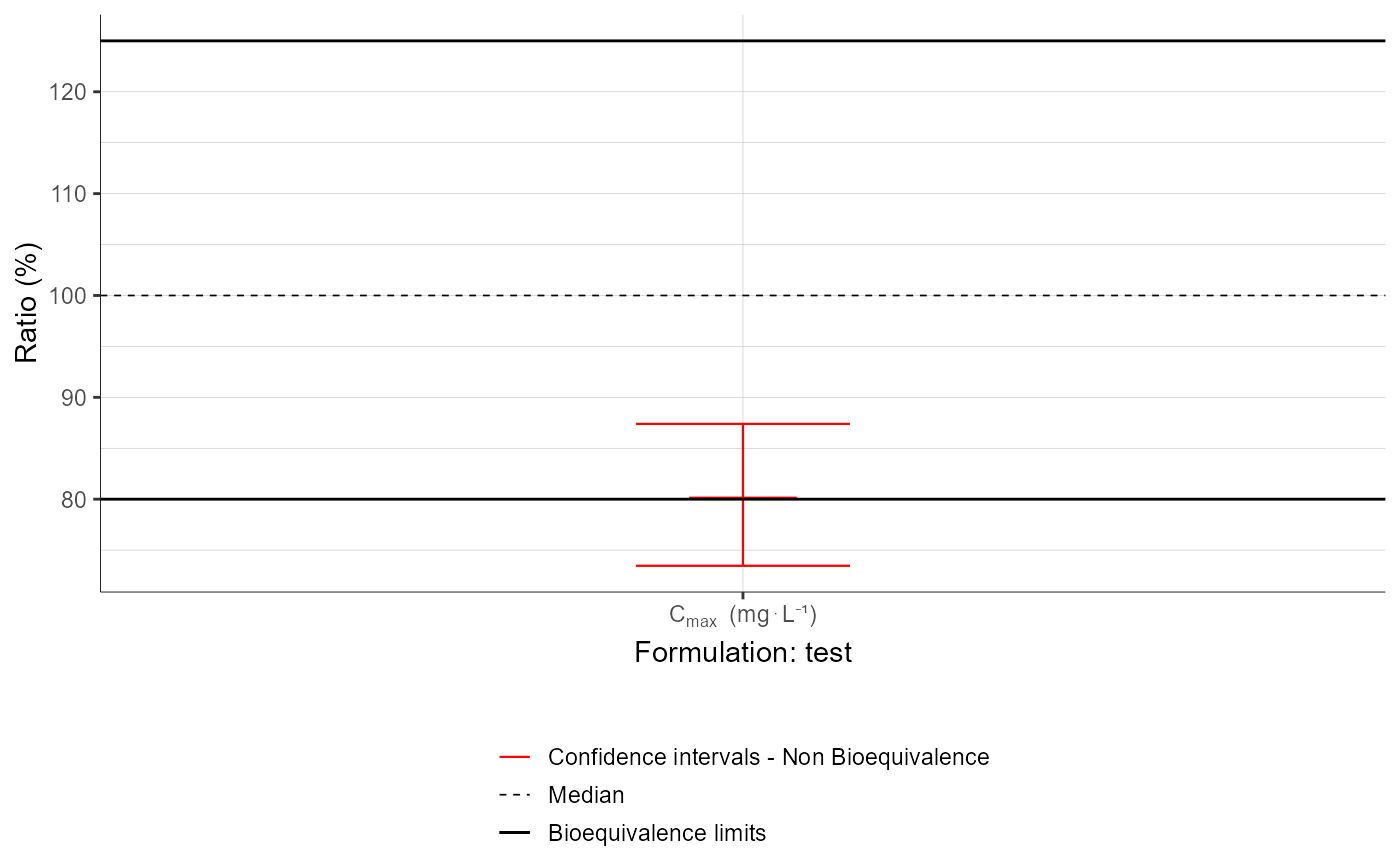
plotBEConfidenceIntervals
[PKanalix] BE Confidence Intervals plot
Plot the individual NCA parameters vs covariates.
Usage
plotBEConfidenceIntervals(
parameters = NULL,
formulations = NULL,
settings = list(),
preferences = NULL
)Arguments
- parameters
vector of bioequivalence parameters to display. (by default the first 4 computed parameters are displayed).
- formulations
vector of test formulations to display. (by default the first 4 test formulations are displayed).
- settings
List with the following settings
median(logical) - If TRUE median is displayed (default TRUE).limits(logical) - If TRUE confidence intervals limits are displayed (default TRUE).legend(logical) add (TRUE) / remove (FALSE) plot legend (default FALSE).grid(logical) add (TRUE) / remove (FALSE) plot grid (default TRUE).ylog(logical) add (TRUE) / remove (FALSE) log scaling on y axis (default TRUE).ylab(character) label on y axis (default Ratio).ncol(integer) number of columns when facet = TRUE (default 4).fontsize(integer) Plot text font size.units(logical) Set units in axis labels (default TRUE).
- preferences
(optional) preferences for plot display, run getPlotPreferences("plotBEConfidenceIntervals") to check available displays.
Value
A ggplot object if one parameter,
A TableGrob object if multiple plots (output of grid.arrange)
See also
Examples
initializeLixoftConnectors(software = "pkanalix")
project <- file.path(getDemoPath(), "2.case_studies/project_Theo_extravasc_SD.pkx")
loadProject(project)
runNCAEstimation()
runBioequivalenceEstimation()
plotBEConfidenceIntervals()

plotBEConfidenceIntervals(parameters = "Cmax",
settings = list(legend = T))
$body