formatData
[Monolix - PKanalix] Adapt and export a data file as a MonolixSuite formatted data set.
Adapt and export a data file as a MonolixSuite formatted data set.
Usage
formatData(
dataFile,
formattedFile,
headerLines = 1,
headers,
linesToExclude = NULL,
observationSettings = NULL,
observations = NULL,
treatmentSettings = NULL,
treatments = NULL,
additionalColumns = NULL,
sheet = NULL
)Arguments
- dataFile
(character) Path to the original data file (csv, xlsx, xlsx, sas7bdat, xpt or txt). Can be absolute or relative to the current working directory.
- formattedFile
(character) Path to the data file that will be exported (must end with the .csv, .txt, .tsv or .xpt extension).
- headerLines
(optional) (integer or vector) Line numbers containing headers (if multiple numbers are given, formatted headers will contain values from all header lines concatenated with the "_" character) - default: 1.
- headers
(list) List of headers or indexes for columns containing information about ID, time, volume (in case of urine data) and sort columns. If the headers are changed by Data Formatting, the original headers should be given.
id(character) - Name of the column distinguishing data from different individuals.time(character) - Name of the column containing observation times (in case of plasma data).sort(character or vector) - Name of the column(s) distinguishing different profiles.start(character) - Name of the column containing urine collection start times (in case of urine data).end(character) - Name of the column containing urine collection end times (in case of urine data).volume(character) - Name of the column containing collected volume of urine samples (in case of urine data).
- linesToExclude
(optional) (integer or vector) Numbers of lines that should be removed from the data set.
- observationSettings
(optional) (list) List containing settings applied when different observation columns are merged into a single column.
distinguishWithObsId(logical) - If TRUE, different observations will be distinguished with the observation ID column (default), otherwise they will be distinguished with occasions.duplicateInformation(logical) - If TRUE, information from undefined columns will be duplicated (default) in the newly created rows.
- observations
(optional) (list) List of lists containing information about different observation types:
header(character) - Name of the column containing observations. If the header is changed by Data Formatting, the original header should be given.censoring(list) - List of lists containing information about different types of censored data (not necessary if there is no censored data):type(character) - Type of censoring, one of "LLOQ", "ULOQ", or "interval".tags(character or vector) - Strings in the observation column indicating that the data is censored (e.g., "BLQ", "LLOQ", ...).limits- Define limits of censored data. If censoring type is "LLOQ" or "ULOQ", the lower and upper limit is defined with one of the following arguments. If censoring type is "interval", the lower and upper limits of the censoring interval are defined with a list of two of the following arguments:as character - The column with the indicated header will be used to define limits.
as double - The value will be used as a lower/upper limit.
as list - Used to give different values for different categories. List needs to be have two arguments:
category(character) - Name of the column containing the category.values(list) - List containing modalities as keys and limit values as values (e.g., list(method1 = 0.06, method2 = 0.1)).
- treatmentSettings
(optional) (list) List containing settings applied to all treatments.
infusionType("rate"|"duration", default = "duration") - Type of values defining infusion.doseIntervalsAsOccasions(default = FALSE) (logical) - If TRUE, occasions will be created for each dose interval.duplicateObservationsAtDoseTimes(default = FALSE) (logical) - If TRUE anddoseIntervalsAsOccasionsis TRUE, doses will duplicate observations if both are at the same time.
- treatments
(optional) (list or character) List that can contain lists with information about different treatments or strings with paths to files that contain treatment information.
Lists with information about different treatments need to have the following elements:times(double or vector) - Times at which the dose is administered (R function seq can be used to define regular treatments).amount(character, double or list) - Administered amount. Can be defined in the same way as censoring limits (through a column name, as a fixed value or as values depending on categories).infusion(character, double or list) - Infusion rate or duration (see the treatmentSettings argument for more information). Can be defined in the same way as censoring limits (through a column name, as a fixed value or as values depending on categories). Does not need to be provided if the drug is not administered through an infusion.admId(character, double or list) - Administration ID. Can be defined in the same way as censoring limits (through a column name, as a fixed value or as values depending on categories). If not provided, default of 1 will be used.repeatCycle(list) - List containing repetition information (does not need to be provided if the treatment is not repeated):duration(double) - Duration of a cycle.number(integer) - Number of repetitions.
Path to files that contain treatment information can be just one path (csv, xlsx, xlsx, sas7bdat, xpt or txt, absolute or relative to the current working directory),
or a list of paths (to combine several treatments):file(character) File path 1file(character) File path 2file(character) etc
or a list of lists with 2 elements to specify for each treatment an xls/xlsx file and sheet in the excel file:
list with
file(character) File path 1sheet[optional] (character): Name of the sheet in first xlsx/xls file.
list with
file(character) File path 2sheet[optional] (character): Name of the sheet in second xlsx/xls file.
etc
- additionalColumns
(optional) (character or vector) Path(s) to the file(s) containing additional columns (needs to have the ID column). Accepted formats are csv, xlsx, xlsx, sas7bdat, xpt or txt. It can be just one path, or a list of paths (to use columns from several external files):
file(character) File path 1file(character) File path 2file(character) etc
or a list of lists with 2 elements to specify an xls/xlsx file and sheet in the excel file:
list with
file(character) File path 1sheet[optional] (character): Name of the sheet in first xlsx/xls file.
list with
file(character) File path 2sheet[optional] (character): Name of the sheet in second xlsx/xls file.
etc
- sheet
[optional] (character): Name of the sheet in xlsx/xls file. If not provided, the first sheet is used.
Details
Data formatting can be performed as in the Data Formatting Tab of Monolix and PKanalix interface. Look at the examples to see how each data formatting demo project could be created with the connectors.
See also
Examples
# example: create a new project with a dataset to format:
initializeLixoftConnectors(software = "pkanalix")
FormattedDataPath = tempfile("formatted_data", fileext = ".csv")
formatData(paste0(getDemoPath(),"/0.data_formatting/data/units_BLQ_tags_data.csv"),
formattedFile = FormattedDataPath,
headerLines = c(1,2),
headers = c(id="ID", time="TIME"),
observations = list(header="CONC",
censoring = list(type="interval", tags = c("BLQ"),
limits=list(0,"LLOQ"))),
treatments = list(times=0, amount=100))
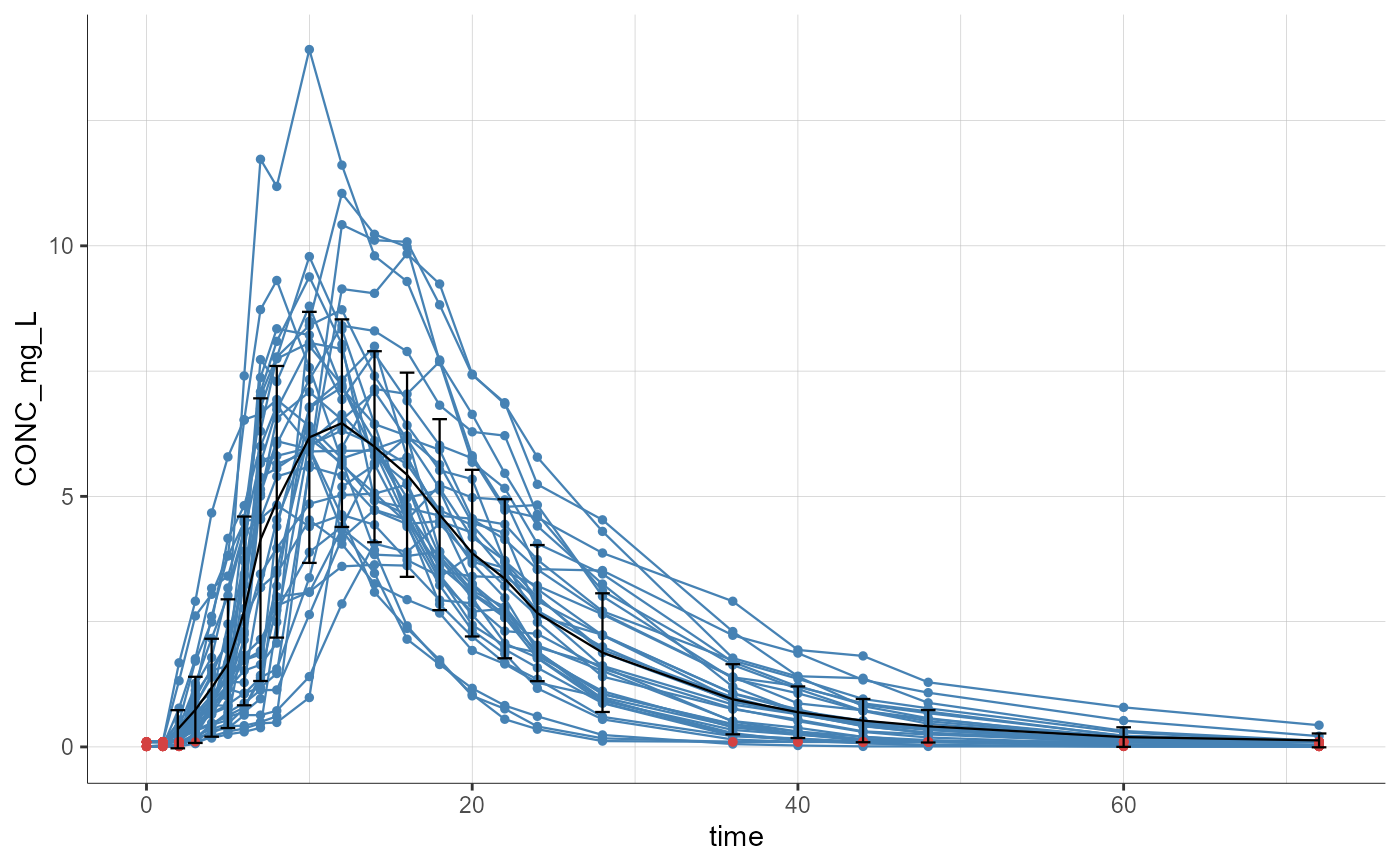
colnames(read.csv(FormattedDataPath)) # to check column names of the generated file and tag them as desired
#> [1] "ID" "TIME_h" "CONC_mg_L" "AGE" "WT" "STUDY"
#> [7] "LLOQ_mg_L" "AMT" "CENS" "LIMIT"
newProject(data = list(dataFile = FormattedDataPath, headerTypes = c("id","time","observation","contcov","contcov","catcov","ignore","amount","cens","limit")))
plotObservedData()
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's linetype values.
# demo merge_occ_ParentMetabolite.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/parent_metabolite_data.csv"),
formattedFile = FormattedDataPath,
headers = c(id="ID", time="TIME"),
observations = list(list(header="PARENT",
censoring = list(type="interval", tags = c("BLQ"), limits=list(0,0.01))),
list(header="METABOLITE")),
observationSettings = list(distinguishWithObsId = FALSE),
treatments = list(times=0, amount="DOSE"))
# demo merge_obsID_ParentMetabolite.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/parent_metabolite_data.csv"),
formattedFile = FormattedDataPath,
headers = c(id="ID", time="TIME"),
observations = list(list(header="PARENT",
censoring = list(type="interval", tags = c("BLQ"), limits=list(0,0.01))),
list(header="METABOLITE")),
treatments = list(times=0, amount="DOSE"))
# demo DoseAndLOQ_byCategory.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/units_BLQ_tags_data.csv"),
formattedFile = FormattedDataPath,
headerLines = c(1,2),
headers = c(id="ID", time="TIME"),
observations = list(header="CONC",
censoring = list(type="interval", tags = c("BLQ"),
limits=list(0,list(category="STUDY",
values=list("SD_400mg"=0.01, "SD_500mg"=0.1, "SD_600mg"=0.1))))),
treatments = list(times=0, amount=list(category="STUDY",
values=list("SD_400mg"=400, "SD_500mg"=500, "SD_600mg"=600))))
# demo DoseAndLOQ_fromData.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/units_BLQ_tags_data.csv"),
formattedFile = FormattedDataPath,
headerLines = c(1,2),
headers = c(id="ID", time="TIME"),
observations = list(header="CONC",
censoring = list(type="interval", tags = c("BLQ"),
limits=list(0,"LLOQ"))),
treatments = list(times=0, amount="STUDY"))
# demo DoseAndLOQ_manual.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/units_multiple_BLQ_tags_data.csv"),
formattedFile = FormattedDataPath,
headerLines = c(1,2),
headers = c(id="ID", time="TIME"),
observations = list(header="CONC",
censoring = list(list(type="interval", tags = c("BLQ1"), limits=list(0,0.06)),
list(type="interval", tags = c("BLQ2"), limits=list(0,0.1)))),
treatments = list(times=0, amount=600))
# demo Urine_LOQinObs.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/urine_LOQinObs_data.csv"),
formattedFile = FormattedDataPath,
headers = c(id="ID", start="START TIME", end="END TIME", volume="VOLUME"),
observations = list(header="CONC",
censoring=list(type="LLOQ", tags="<LOQ=1>", limits="CONC")),
treatments = list(paste0(getDemoPath(),"/0.data_formatting/data/urine_data_doses.csv")))
# demo CreateOcc_AdmIdbyCategory.pkx
formatData(paste0(getDemoPath(),"/0.data_formatting/data/two_formulations_data.csv"),
formattedFile = FormattedDataPath,
linesToExclude = 1, headerLines = c(2,3),
headers = c(id="ID", time="TIME", sort="FORM"),
observations = list(header="CONC",
censoring=list(type="LLOQ", tags="BLQ", limits=0.06)),
treatments = list(times=0, amount=600, admId=list(category="FORM", values=list("ref"=1,"test"=2))))
# MONOLIX EXAMPLES
initializeLixoftConnectors(software = "monolix")
FormattedDataPath = tempfile("formatted_data")
# demo doseIntervals_as_Occ.mlxtran
formatData(paste0(getDemoPath(),"/0.data_formatting/data/data_multidose.csv"),
formattedFile = FormattedDataPath,
headers = c(id="ID", time="TIME"),
observations = list(header="CONC"),
treatments = list(times=seq(0,by=12,length=7), amount=40),
treatmentSettings = list(doseIntervalsAsOccasions = TRUE))
# demo warfarin_PKPDseq_project.mlxtran
formatData(paste0(getDemoPath(),"/0.data_formatting/data/warfarin_data.csv"),
formattedFile = FormattedDataPath,
headers = c(id="id", time="time"),
additionalColumns = paste0(getDemoPath(),"/0.data_formatting/data/warfarinPK_regressors.txt"))
