defineTreatmentElement
[Simulx] Define treatment element
Define or edit a treatment element.
Usage
defineTreatmentElement(name, element)Arguments
- name
(character) Element name.
- element
(list) List with the treatment settings:
data(mandatory)(data frame OR path to external file (csv, xlsx, xlsx, sas7bdat, xpt or txt) OR list to select the sheet of an excel file: list( file = path_to_excel, sheet = sheet_name ) Column headers: [only data frame] for a regular treatment common to all ids:
occ(optional, if common occasion structure, same header as in the occasion element),start,interval,nbDoses,amount[only data frame] for a manual treatment common to all ids:
occ(optional, if common occasion structure, same header as in the occasion element),time,amount,washout(optional, to add a washout just before the dose, otherwise washout = FALSE by default)[only external file] for a manual treatment common or specific to each id:
id(optional),occ(optional),time,amount,washout(optional)[data frame or external] in case of infusion:
tInf(duration) ORrate(mutually exclusive).
admID(optional)(integer) same as integer in the model as administration type probaMissDose(optional)(double) probability to miss a dose (number in [0 1]) repeats(optional)(vector) to repeat the specified treatment after a specific duration. Elements: cycleDurationduration after which the treatment will be repeated (can be longer than the treatment duration)
NumberOfRepetitionsnumber of times the treatment will be repeated
scale(optional)(list) to scale the dose amount by a covariate. The scaled amount will be administered instead of amount. covariate(character) covariate to use for scaling (same name as in the [COVARIATE] block of the model)
intercept(double, required for continuous covariate): intercept to use in the scaling formula: scaledAmount = amount*cov + intercept
modalities(list, required for categorical covariate): list of lists with, for each modality, the name of the modality (eg "male"), the factor, and the intercept to use in the scaling formula: scaledAmount = [cov = modality]*factor*amt + intercept (no scaling if factor =1 and intercept = 0)
scaleDuration(optional, logical) if TRUE (default), infusion duration will be scaled, otherwise it will be rate
Details
Treatment elements are defined and used for simulation as in Simulx GUI. As for other elements, the treatment elements can be defined or created at import, and they are saved with the Simulx project if calling saveProject. Once an output element is defined, it needs to be added to a simulation group with to be used in simulation. Several treatment elements can be added to the same simulation group and they will be both administered to every individual in the group.setGroupElement
To define a treatment element, in addition to the element name, you need to provide a list with at list one field "data" containing the dose information. The field data can be specified with a data frame or an external file (csv, xlsx, xlsx, sas7bdat, xpt or txt).
To define a regular treatment, common to all individuals, you can use a data frame, with column headers start, interval, nbDoses and amount. You can include an optional column tInf or rate to define an infusion. If the project has a common occasion structure (i.e. same occasions for all individuals), this data frame can contain a column occasion to define the regular treatment occasion-wise. The occasion headers must correspond to the occasion names defined in the occasion element.
To define a treatment by giving a specific list of times and amounts, both data frames and external files (csv, xlsx, xlsx, sas7bdat, xpt or txt) can be used, with a column time. They can contain columns id and occasions (optional). The occasion headers must correspond to the occasion names defined in the occasion element.
Data frames can be used only to define output elements of type 'common', i.e the same for all individuals (potentially occasion-wise). If you want to define subject-specific treatment elements, you have to use an external file with an "id" column.
An external file can be used in all cases (common or subject-specific). It can contain a column id (optional) in addition to occasions (optional), and should contain one column time (mandatory) and one column amount (mandatory). When id and occasion columns are present, then they must be the first columns. When the id column is not present, the covariate is considered common.
Note
To define a regular schedule, it is advised to use a regular treatment without repeats, rather than a manual treatment with repeats. Repeats are useful to create more complex schedules in addition to a manual or regular definition, such as dosing regimen 3 weeks ON, 1 week OFF.
To see the impact of a treatment until the end of a dosing regimen, you should set an output element that spans the duration of the treatment to the same simulation group.
See also
Examples
if (FALSE) {
defineTreatmentElement(name = "name", element = list(data = "file/path"))
defineTreatmentElement(name = "name", element = list(data = list(file = "file/path", sheet = "sheetname")))
defineTreatmentElement(name = "name", element = list(probaMissDose=0, admID=1, repeats=c(cycleDuration = 1, NumberOfRepetitions=12), data=data.frame(time=c(1,2), amount=c(1,2), tInf=c(0, 1), washout=c(TRUE, FALSE))))
defineTreatmentElement(name = "name", element = list(probaMissDose=0, admID=1, repeats=c(cycleDuration = 1, NumberOfRepetitions=12), data=data.frame(time=c(10,10,10,10), amount=c(10,20,30,40), occ1=c(1,1,2,2), occ2=c(1,2,1,2))))
defineTreatmentElement(name = "name", element = list(probaMissDose=0, admID=1, repeats=c(cycleDuration = 1, NumberOfRepetitions=12), data=data.frame(start=1, interval=2, nbDoses=10, amount=1)))
defineTreatmentElement(name = "name", element = list(admID=1, scale=list(covariate="age", intercept=12), data=data.frame(start=1, interval=2, nbDoses=10, amount=1)))
defineTreatmentElement(name = "name", element = list(admID=1, scale=list(covariate="sex", modalities=list(list(name="0", factor=1, intercept=10), list(name="1", factor=1.5, intercept=10))), data=data.frame(start=1, interval=2, nbDoses=10, amount=1)))
}
##### Working example with treatment combinations #####
# In this demo, the first group receives only the chemotherapy, while the second group receives both the chemotherapy and the anti-angiogenic therapy.
# Note that the chemotherapy treatment uses adm=1 to be applied to compartment 1 via the macro iv(adm=1, cmt=1) in the model representing the concentration of the chemo drug.
# The anti-angiogenic treatment is defined with adm=2 which is applied via the macro iv(adm=2, cmt=2) to compartment 2 representing the concentration of anti-angiogenic drug.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_combinations.smlx"))
# to see how the structural model is defined:
file.show(getStructuralModel())
defineTreatmentElement(name = "Chemotherapy", element = list(data=data.frame(start=10, interval=14, nbDoses=10, amount=1)))
defineTreatmentElement(name = "AntiAngionenic_treatment", element = list(admID = 2, data=data.frame(start=10, interval=7, nbDoses=20, amount=1)))
setGroupElement("simulationGroup1","Chemotherapy")
setGroupElement("simulationGroup2",c("Chemotherapy","AntiAngionenic_treatment"))
runSimulation()
# use ggplot or export to Monolix/PKanalix to plot trajectories
exportProject(settings = list(targetSoftware = "monolix"),force = TRUE)
if (FALSE) {
plotObservedData( settings = list(dots = FALSE, ylab = "Target Occupancy", legend = TRUE), stratify = list(colorGroup = list(name = "group")), preferences = list(obs = list(lineWidth = 0.5)))
}
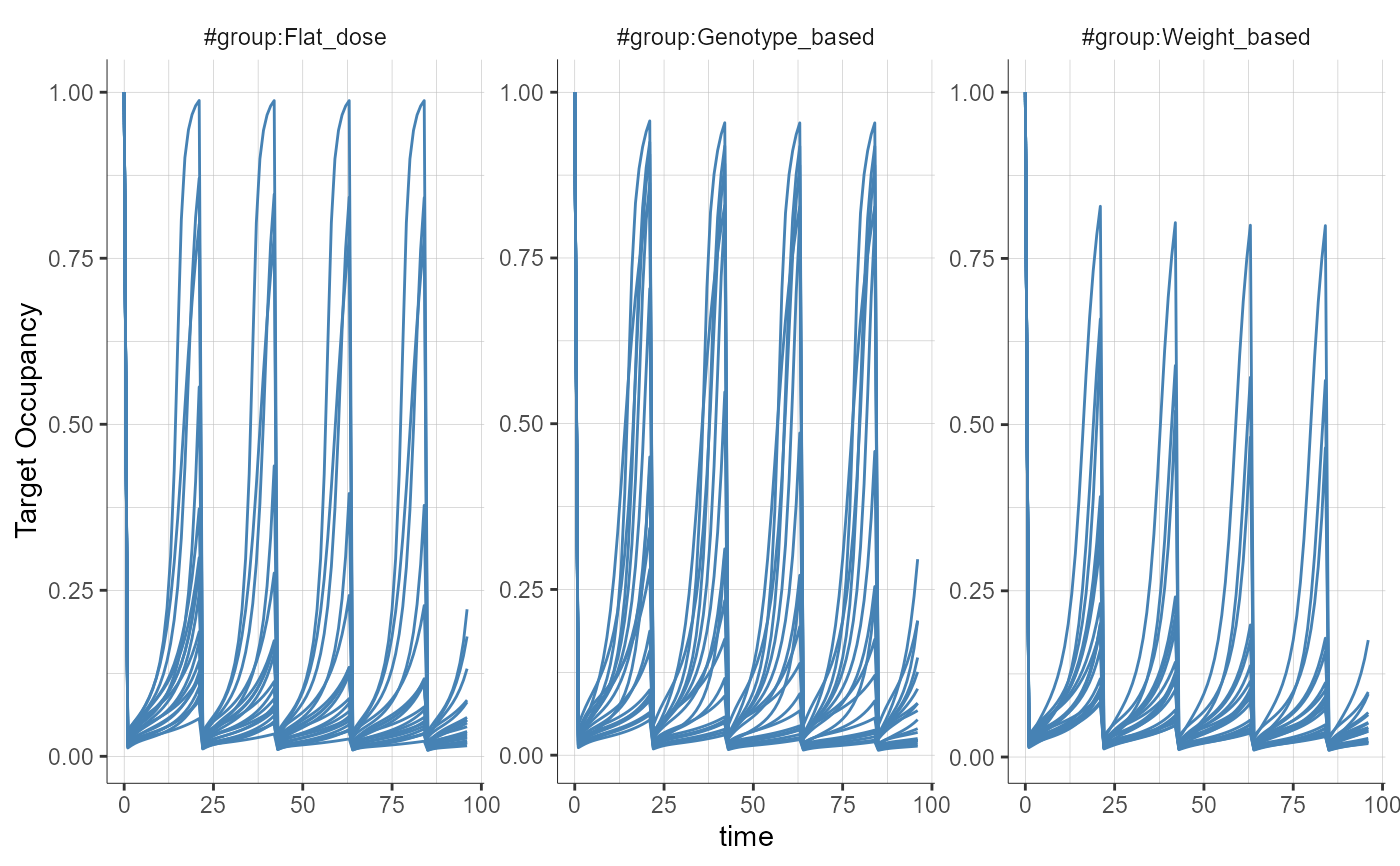
##### Working example with a treatment scaled by weight and based on genotype #####
# In this demo, a weight-based dose is defined by indicating the dose per unit weight in the amount box (14 nmol/kg) and using the "Scale amount by a covariate" option with "Weight" selected as covariate.
# The "intercept" could be used to define a offset common to all weights (e.g 14nmol/kg + 10nmol).
# When an infusion duration or rate has been defined, the user can choose if the infusion duration or the infusion rate is scaled by the covariate.
# For categorical covariates, such as the genotype, a scaling factor and an intercept can be defined for each category.
# In this demo, the scaling for Homozygous is 1 meaning that they receive the dose defined in the amount box.
# For heterozygous, the scaling is 0.8, meaning that they receive 0.8 times the amount in the amount box.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_weight_and_genotype_based.smlx"))
defineTreatmentElement(name = "14nmolPerKg", element = list(data=data.frame(start=0, interval=21, nbDoses=5, amount=14, tInf = 0.208), scale=list(covariate="Weight", intercept = 0, scaleDuration = FALSE)))
defineTreatmentElement(name = "1000nmol", element = list(data=data.frame(start=0, interval=21, nbDoses=5, amount=1000, tInf = 0.208)))
defineTreatmentElement(name = "1000nmolHomo_800nmolHetero", element = list(data=data.frame(start=0, interval=21, nbDoses=5, amount=1000, tInf = 0.208), scale=list(covariate="Genotype", modalities=list(list(name="Homozygous", factor=1, intercept=0), list(name="Heterozygous", factor=0.8, intercept=0)), scaleDuration = FALSE)))
setGroupElement("Weight_based","14nmolPerKg")
setGroupSize("Weight_based",20)
setGroupElement("Flat_dose","1000nmol")
setGroupSize("Flat_dose",20)
setGroupElement("Genotype_based","1000nmolHomo_800nmolHetero")
setGroupSize("Genotype_based",20)
runSimulation()
# use ggplot or export to Monolix/PKanalix to plot trajectories
exportProject(settings = list(targetSoftware = "monolix"),force = TRUE)
plotObservedData(obsName = "yTO", settings = list(dots = FALSE, ylab = "Target Occupancy"), stratify = list(splitGroup = list(name = "group")), preferences = list(obs = list(lineWidth = 0.5)))
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's linetype values.
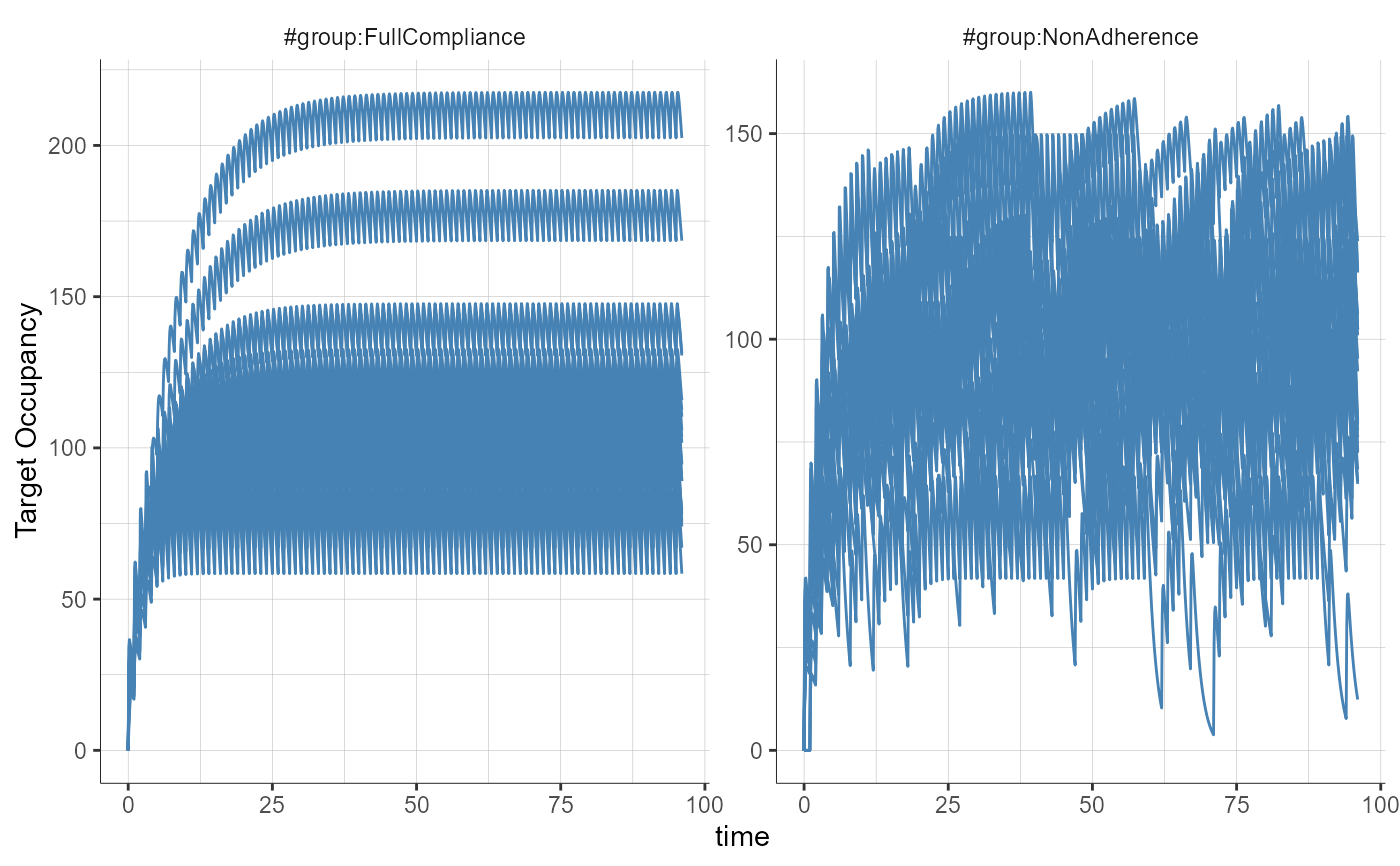
##### Working example with a probability to miss a dose #####
# In this demo, the second treatment is defined with a probability to miss a dose of 0.1, meaning that on average 10% of the doses will not be taken. The missed doses are random.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_non_adherence.smlx"))
defineTreatmentElement(name = "OncePerDay_full_compliance", element = list(data=data.frame(start=0, interval=1, nbDoses=112, amount=100)))
defineTreatmentElement(name = "OncePerDay_partial_compliance", element = list(data=data.frame(start=0, interval=1, nbDoses=112, amount=100),probaMissDose = 0.1))
setGroupElement("simulationGroup1","OncePerDay_full_compliance")
renameGroup("simulationGroup1","FullCompliance")
setGroupElement("simulationGroup2","OncePerDay_partial_compliance")
renameGroup("simulationGroup2","NonAdherence")
setGroupSize("FullCompliance",20)
setGroupSize("NonAdherence",20)
runSimulation()
# use ggplot or export to Monolix/PKanalix to plot trajectories
exportProject(settings = list(targetSoftware = "monolix"),force = TRUE)
plotObservedData( settings = list(dots = FALSE, ylab = "Target Occupancy"), stratify = list(splitGroup = list(name = "group")), preferences = list(obs = list(lineWidth = 0.5)))
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's linetype values.
##### Working example with an external file #####
# Demo: use an external file to define a dose by age group: 1-2 years 12.5 mg, 3-6 years 18.75 mg and 7-15 years 25 mg.
# The age also appears as covariate in the model and the covariate element is defined via an external file.
# To make sure the covariates are sampled from the covariate external file and the doses sampled from the treatment external file are consistent (i.e correspond to the same id and thus the same age), the option "shared id" is selected between covariate and treatment elements.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_external_byAgeGroup.smlx"))
tableAge = getCovariateElements()$External_AGE_values$data
AmtByAgeGroups = (tableAge$AGE < 3)*12.5 + ((tableAge$AGE >=3) & (tableAge$AGE < 7))*18.75 + (tableAge$AGE >= 7)*25
Nid = length(AmtByAgeGroups)
dataAmtByAgeGroups = data.frame(id = tableAge$ID, time = rep(0,Nid), amount = AmtByAgeGroups)
file_name <- tempfile("trt", fileext = ".csv")
write.csv(dataAmtByAgeGroups, file_name, row.names = FALSE)
defineTreatmentElement(name = "doseByAgeGroup", element = list(data = file_name))
setGroupElement("simulationGroup1",c("doseByAgeGroup","External_AGE_values","regularCc"))
setSharedIds(c("covariate", "treatment"))
runSimulation()
# use ggplot or export to Monolix/PKanalix to plot trajectories
exportProject(settings = list(targetSoftware = "monolix"),force = TRUE)
plotObservedData(settings = list(dots = FALSE, ylab = "Cc",legend = TRUE, ylim = c(0,13)), stratify = list(splitGroup = list(name = "AGE", breaks = c(2,7)), colorGroup = list(name = "ID")), preferences = list(obs = list(lineWidth = 0.5)))
#> [ERROR] - Invalid stratify inputs:
#> - Invalid covariate input for color: 'ID'
#> [ERROR] Error during observed data (outputplot) chart creation:
#> - Invalid stratify inputs:
#> - Invalid covariate input for color: 'ID'
##### Working example with washout #####
# In this demo, two different formulations are given.
# The reference formulation is given at time zero.
# The test formulation is given after a long washout period.
# In order not to simulate this washout period, the test dose is defined at time 48 and a washout is applied just before the test dose to reset the model to its initial state.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_washout.smlx"))
defineTreatmentElement(name = "ReferenceFormulation_atTime0", element = list(data=data.frame(time=0, amount=600)))
defineTreatmentElement(name = "TestFormulation_atTime48", element = list(admID = 2, data=data.frame(time=0, amount=600, washout = TRUE)))
setGroupElement("simulationGroup1",c("ReferenceFormulation_atTime0","ReferenceFormulation_atTime0"))
##### Working example with a regular treatment and repeats #####
# The "repeat" option allows to repeat a base pattern several times with a defined periodicity.
# In this demo, the first treatment is defined as one dose per day during 112 days.
# The second treatment is defined as one dose per day during 14 days and this is repeated every 28 days leading to a 2 weeks on / 2 weeks off pattern.
initializeLixoftConnectors("simulx")
loadProject(paste0(getDemoPath(), "/3.definition/3.1.treatments/treatment_regular_cycles.smlx"))
defineTreatmentElement(name = "OncePerDay_4weeksOn", element = list(data=data.frame(start=0, interval=1, nbDoses=112, amount=100)))
defineTreatmentElement(name = "OncePerDay_2weeksOn2weeksOff", element = list(repeats=c(cycleDuration = 28, NumberOfRepetitions=4), data=data.frame(start=0, interval=1, nbDoses=14, amount=100)))
setGroupElement("simulationGroup1","OncePerDay_4weeksOn")
renameGroup("simulationGroup1","4weeksOn")
setGroupElement("simulationGroup2","OncePerDay_2weeksOn2weeksOff")
renameGroup("simulationGroup2","2weeksOn2weeksOff")
runSimulation()
# use ggplot or export to Monolix/PKanalix to plot trajectories
exportProject(settings = list(targetSoftware = "monolix"),force = TRUE)
plotObservedData(settings = list(dots = FALSE, ylab = "Cc",legend = TRUE), stratify = list(color = list(name = "group")), preferences = list(obs = list(lineWidth = 0.5)))
#> [ERROR] Unexpected type encountered for 'state' : 'color' field must be a vector of string.

$body