Friberg model for myelosupression
The Friberg model is a semi-mechanistic pharmacodynamic model describing chemotherapy-induced myelosuppression. It has been originally developed to describe leukocyte and neutrophil data after administration of docetaxel, paclitaxel, etoposide, DMDC and irinotecan and vinflunine in Friberg et al. (2002), and has been reused in many other publications since.
Model description
The model consists of a proliferating compartment that is sensitive to drugs, three transit compartments that represent maturation, and a compartment of circulating blood cells. The parameterization is parsimonious (initial circulating concentration, mean transit time and a feedback parameter), well suited for sparse data. The drug effect can be implemented as a linear function or with an Emax model, as shown below.
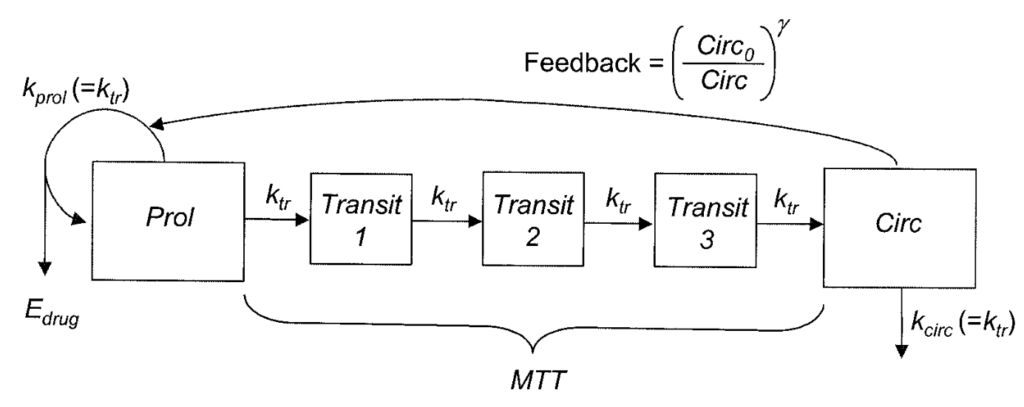
The PK part of the model is in this example a (ka,V,Cl) model but can be replaced by any other PK model. PK parameters can either be estimated (as below) or read from the data set (uncomment the three lines below the input line to consider the PK parameters as regressors).
Mlxtran model code
DESCRIPTION:
PD = Friberg et al (2002) 'Model of Chemotherapy-Induced Myelosuppression with Parameter Consistency Across Drugs', Journal of Clinical Oncology 20:4713-4721
[LONGITUDINAL]
input = {ka, V, Cl, Circ0, MTT, gam, Emax, EC50}
; ka = {use=regressor}
; V = {use=regressor}
; Cl = {use=regressor}
EQUATION:
odeType=stiff
; PK model
Cc = pkmodel(ka, V, Cl)
; parameter transformations
n = 3
ktr = (n+1)/MTT
kprol = ktr
kcirc = ktr
; drug effect
Edrug = Emax * Cc/(EC50 + Cc)
; initial values
t_0 = 0
Prol_0 = Circ0
T1_0 = Circ0
T2_0 = Circ0
T3_0 = Circ0
Circ_0 = Circ0
;ODEs for the response
ddt_Prol = kprol*Prol*(1-Edrug)*(Circ0/Circ)^gam - ktr*Prol
ddt_T1 = ktr*Prol - ktr*T1
ddt_T2= ktr*T1 - ktr*T2
ddt_T3 = ktr*T2 - ktr*T3
ddt_Circ = ktr*T3 - kcirc*Circ
OUTPUT:
output = {Cc, Circ}In the Monolix GUI, a lognormal distribution can be chosen for Circ0, MTT, gam and EC50, and a logit distribution for Emax to ensure that (1-Edrug) stays always positive. It is common to remove the random effects on gam.
