Antibody–drug conjugates or ADCs are a class of drugs designed as a targeted therapy for treating cancer. They are complex molecules composed of an antibody that targets a specific tumor antigen, linked to a biologically active cytotoxic payload or drug.
On this page we show an example of population PK model for an ADC drug published in:
Li, H., Han, T.H., Hunder, N.N., Jang, G. and Zhao, B. (2017), Population Pharmacokinetics of Brentuximab Vedotin in Patients With CD30-Expressing Hematologic Malignancies. The Journal of Clinical Pharmacology, 57: 1148-1158. https://doi.org/10.1002/jcph.920
In the publication the model is written for NONMEN, here we have translated it in Mlxtran for use with the MonolixSuite.
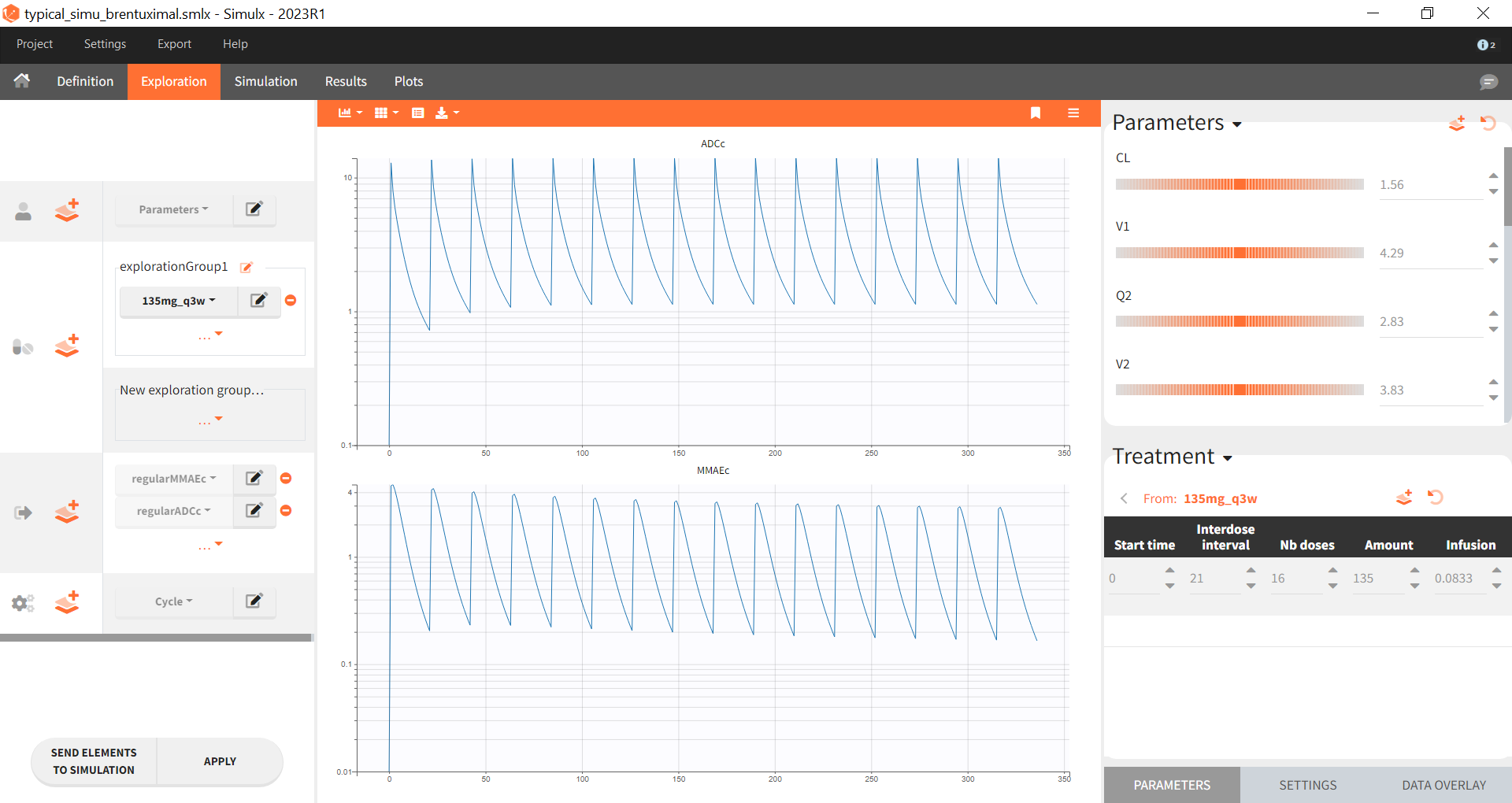
The model describes the PK of brentuximab vedotin, a CD30-directed antibody-drug conjugate (ADC), which is approved for treating certain patients with CD30-expressing hematologic
malignancies. Its primary mechanism of action is the targeted delivery of a microtubule-disrupting agent, monomethyl auristatin E (MMAE), to CD30-expressing cells. In the model considers intravenous administration of brentuximab vedotin and characterizes also the PK of unconjugated MMAE.
The following figure from the publication describes the model:
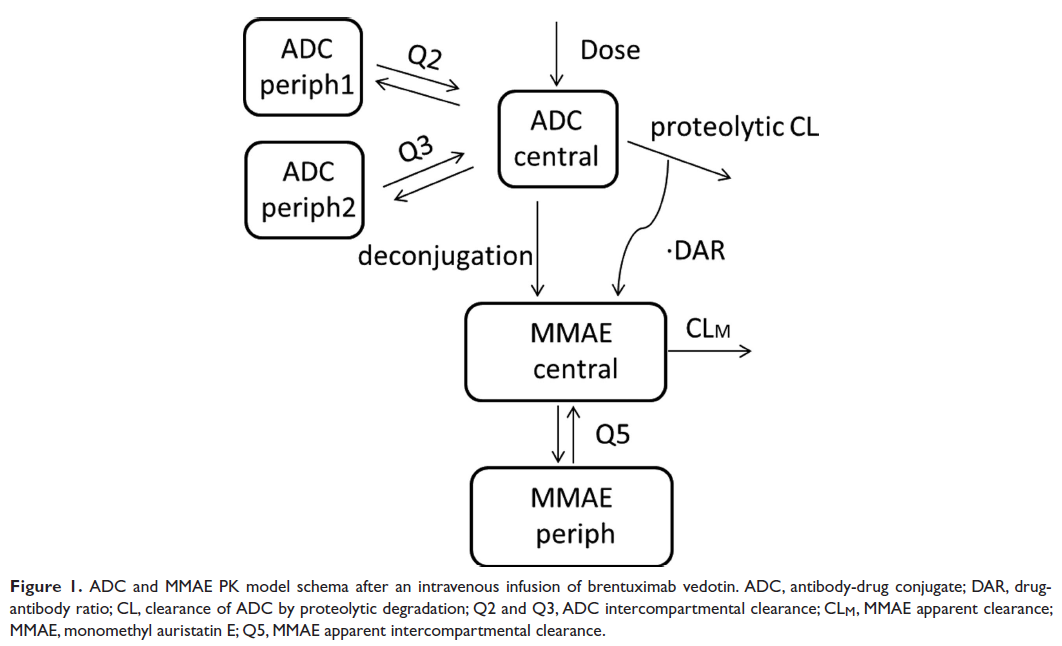
Mlxtran code for the ADC and MMAE population PK model
The full Mlxtran code is given below:
DESCRIPTION:
ADC–MMAE model describing ADC and MMAE concentrations following intravenous administration of brentuximab vedotin.
Published in:
Li, H., Han, T.H., Hunder, N.N., Jang, G. and Zhao, B. (2017), Population Pharmacokinetics of Brentuximab Vedotin in Patients With CD30-Expressing Hematologic Malignancies. The Journal of Clinical Pharmacology, 57: 1148-1158. https://doi.org/10.1002/jcph.920
[LONGITUDINAL]
input={CL, V1, Q2, V2, Q3, V3, CLM, V4, Q5, V5, CYCLE, FM, DAR0, ALPHA, BETA}
CYCLE = {use = regressor}
PK:
MW_ADC = 153000 ; molecular weight of ADC [g/mol]
MW_MMAE = 718 ; molecular weight of MMAE [g/mol]
depot(target=A1, p=1e3/MW_ADC) ;from mg to nmol
EQUATION:
odeType=stiff
K10=CL/V1
K12=Q2/V1
K21=Q2/V2
K13=Q3/V1
K31=Q3/V3
K40=CLM/V4
K45=Q5/V4
K54=Q5/V5
FMC = CYCLE^FM ; FRACTION OF ADC CONVERTS TO MMAE BY CYCLE
DAR=DAR0*(ALPHA + (1-ALPHA)*exp(-BETA*(t-tDose)))
ddt_A1 = - (K10 + K12 + K13)*A1 + K21*A2+ K31*A3 ; ADC CENTRAL COMPT
ddt_A2 = K12*A1 - K21*A2 ; ADC PERI COMPT -1
ddt_A3 = K13*A1 - K31*A3 ; ADC PERI COMPT -2
ddt_A4 = K10*A1*DAR*FMC + A1*(DAR-DAR0*ALPHA)*BETA-K40*A4-K45*A4 + K54*A5 ; MMAE CENTRAL COMPT
ddt_A5 = K45*A4 - K54*A5 ;MMAE PERI COMPT
; Proteolytic pathway
ddt_A6 = K10*A1*DAR
; deconjugation pathway
ddt_A7 = A1*(DAR-DAR0*ALPHA)*BETA
ADCc_pmol_mL = A1/V1 ; in nmol/L
MMAEc_pmol_mL = A4/V4 ; in nmol/L
ADCc = ADCc_pmol_mL*(MW_ADC/1e3) ; from nmol/L to ug/mL
MMAEc = MMAEc_pmol_mL*(MW_MMAE/1) ; from nmol/L to ng/mL
OUTPUT:
output = {ADCc, MMAEc}
The model has two outputs for ADC concentration and MMAE concentration.
The average drug load (or drug-antibody ratio DAR) was not measured but was assumed to decrease exponentially after each dose. In Mlxtran, the equation for DAR uses the keyword tDose that corresponds to the time of the last dose.
CYCLE is a time-varying regressor indicating the number of the last dose cycle. The fraction of ADC converted to MMAE by cycle varies with FMC = CYCLE^FM.
The model uses the molecular weights of ADC and MMAE to convert the dose amount from mg to nmol and the output concentrations from nmol/L to ug/mL or ng/mL.
Typical simulations of the model in Simulx
This Simulx project (download) uses the brentuximal model and the population estimates from the paper to simulate a typical prediction:
(note that the link points to a zip file, so the download might be blocked by your browser, in that case you need to right-click > Save link as > Keep)