Building a simulation scenario
Simulx allows to interactively build a simulation scenario using simulation elements defined in the “Definition” tab and additional options available in the interface. It is done in the Simulations tab – Simulation section – which contains:
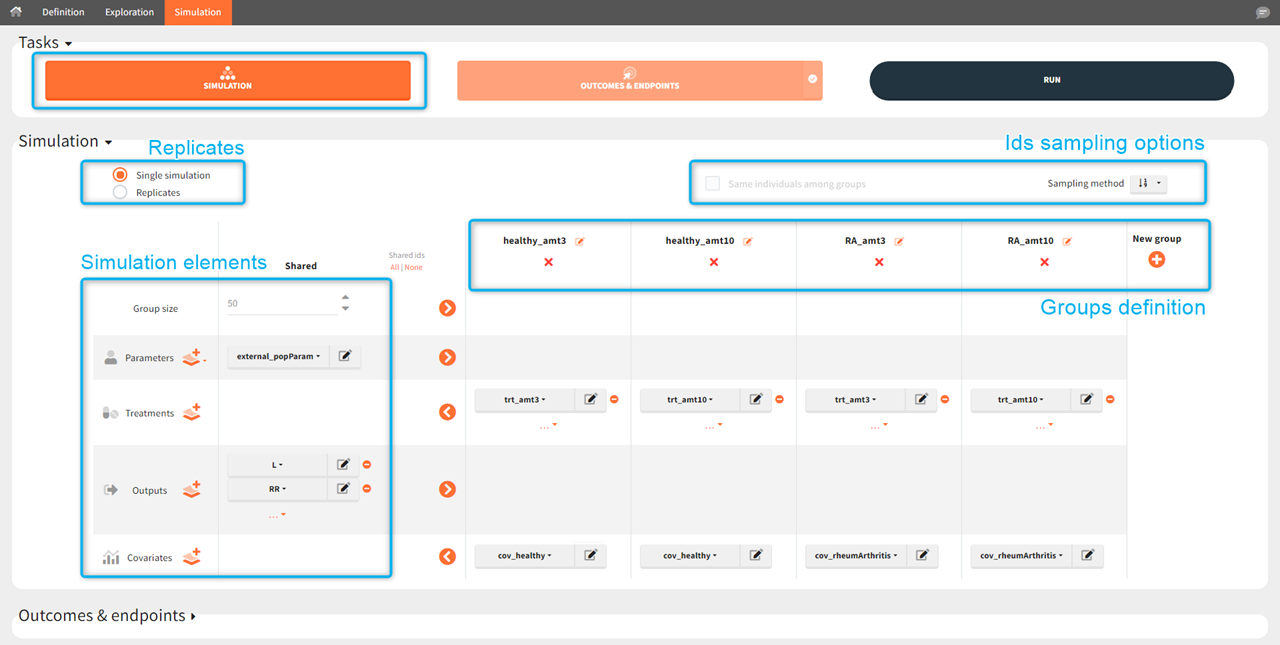
List of simulation elements, which can be shared between groups or set as group specific.
Flexible group definition with drop-down menus for selecting group specific elements and with editable group names.
Option to run a single simulation or replicates.
Different sampling options such as using subjects with the same individual parameters or drawing individuals from table type elements with different methods.
To run a scenario, just click on the button SIMULATION in the tasks section. When importing a Monolix project, Simulx automatically sets the scenario re-simulating the Monolix project.
Simulation scenario elements
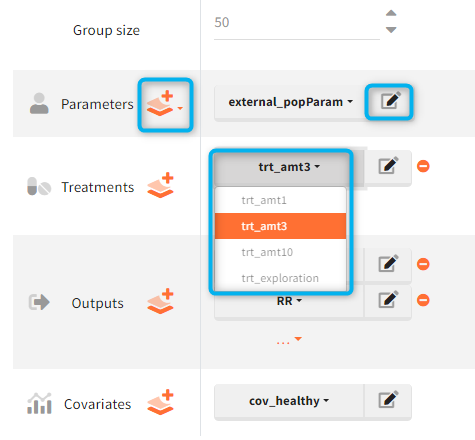
List of simulation elements contains: group size, parameters, treatments, outputs and, if used in a model, covariates and regressors. To select an element from already defined elements, use a drop-down menu available after clicking on the currently selected element. To edit a selected element, click on the icon ![]() . To create a new element, click on the “plus” icon
. To create a new element, click on the “plus” icon ![]() to open a “New element” pop-up window in the Definition tab.
to open a “New element” pop-up window in the Definition tab.
Mandatory elements
Group size corresponds to the number of individuals in a group.
Parameters can be chosen from POP. PARAM. or INDIV. PARAM. elements in the Definition. When Individual parameters are selected, then the covariate element disappears and new section to set the error model parameters becomes available.

Remaining parameters are all parameters that appear in the structural model (in the input line of [LONGITUDINAL]) and are neither individual parameters not regressors. They are typically error model parameters. These error model parameters will impact the simulation only if a noisy observation (from the DEFINITION section of the [LONGITUDINAL] block) is set as output element (instead of a smooth prediction in OUTPUT or variable in EQUATION).
Outputs correspond to one or more variables to be computed and displayed in plots and results. When importing a project from Monolix, by default all observation outputs (if defined in a model) are automatically selected, otherwise only the last defined model output.
Covariates are available in two situations: selected treatments use a covariate scaling or individual parameters model uses covariates and population parameters are selected as an element in the Parameters.
Regressor is available only if it is used is a model.
Optional elements
Treatment can be one element or a combination of several treatment elements.
Occasions are optional even if defined in a model, but are mandatory if any occasion-dependent element is selected in a scenario.
Simulx automatically assigns default elements: the last one from the definition list of each category or specific default “mlx_” types after importing a Monolix, see here for details. It is not possible to remove in the Definition tab an element currently used in a simulation. However, it might not be possible to run a simulation if any of the mandatory elements was removes. For instance, deleting occasions element removes automatically all occasion-dependent elements even if used in a simulation.
Simulation scenario with groups
Groups allows to build a scenario with population having different characteristics, such as parameters, regressors, covariates, different treatments and/or different simulation outputs. Moreover, groups can have different number of individuals to compare clinical trial of different sizes.
To create a new group click on the “plus” button, use the arrows to move elements from “shared” part to group section (or vice versa) and select specific element for each group. Each simulation element can be shared between groups or can be a group specific.
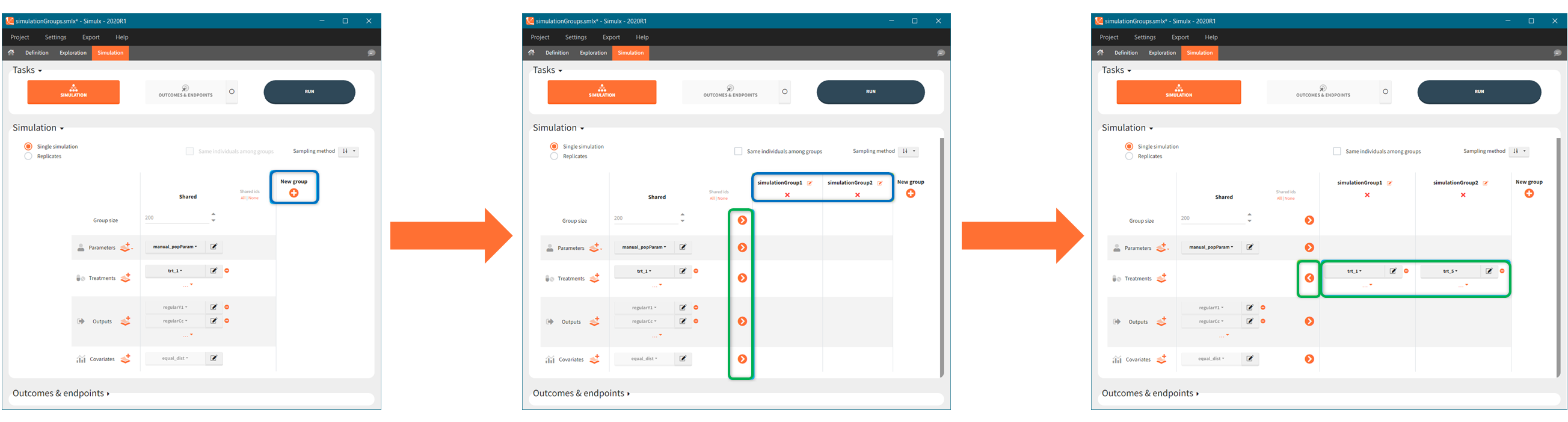
To edit a group name click ![]() next to it. In the Plots tab, figures for different groups appear in the same order as they are defined in the Simulation tab, not in the alphabetical order. To remove a group click “x” under the group name.
next to it. In the Plots tab, figures for different groups appear in the same order as they are defined in the Simulation tab, not in the alphabetical order. To remove a group click “x” under the group name.
Each new group is by default a copy of the previous one (eg. group 3 is a copy of group 2).
Population and individual parameters can not be used in different groups simultaneously.
Examples:
[Demo 5.simulation: simulationGroups_treatment]: Simulation of three groups with three different oral dose levels: low (trt_1), medium (trt_5) and high dose (trt_10). Groups sizes, parameters, covariates and outputs are shared (the same) between the groups, while treatment is group specific.
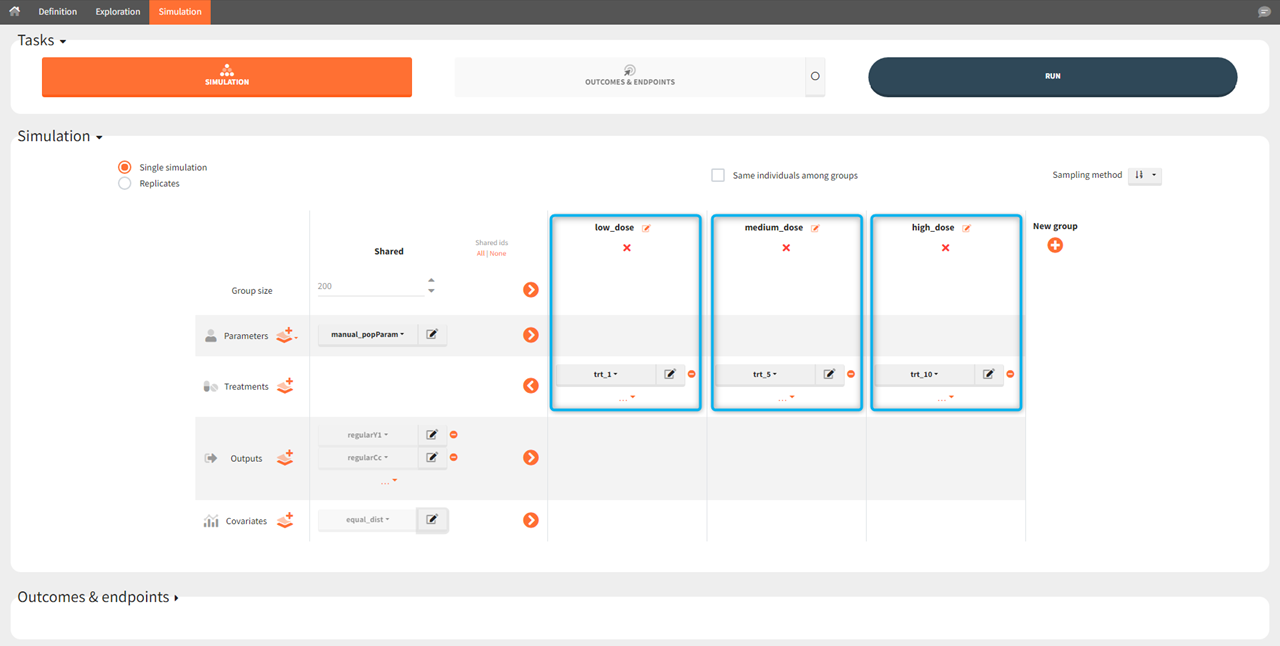
[Demo 5.simulation: simulationGroups_ethnicGroups]: Simulation of two groups with different ethnicity, which affects the bioavailability. In this case, “covariates” simulation element is group specific with: “caucasian” modality for the first group and “asian” for the second group.

Simulation scenario with replicates
By default, Simulx simulates one clinical trial. When the option “replicates” is selected, then Simulx simulate the scenario several times.
If population parameters are vectors (user-defined or imported from a Monolix project), then all replicates have the same population parameters.
If population parameters are of a distribution type, then Simulx samples one set of population parameters for each replicate.
If population parameters are given as an external table with several population parameters sets, for instance from the simpopmlx function, then each replicate uses one set of population parameters with the order of the appearance in the table.
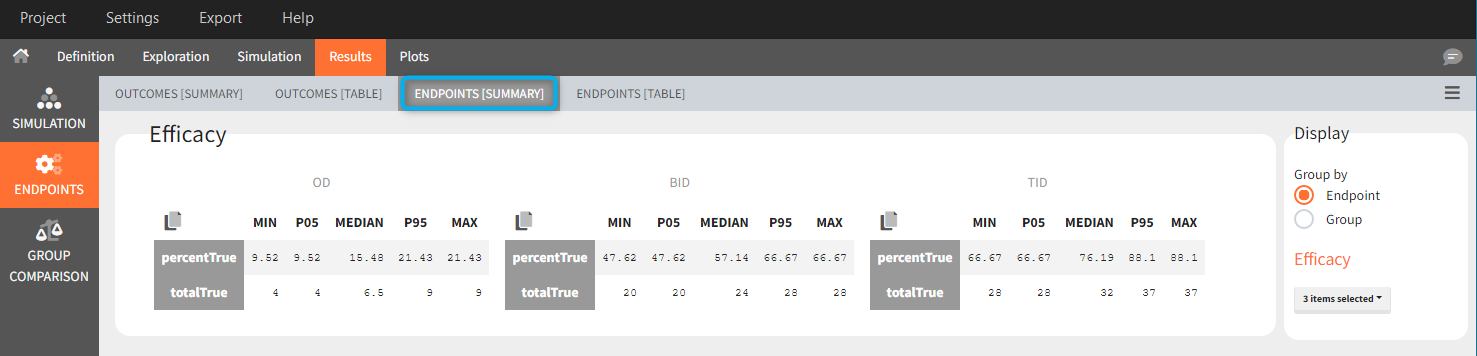
Tables in the results tab display information for all replicates.
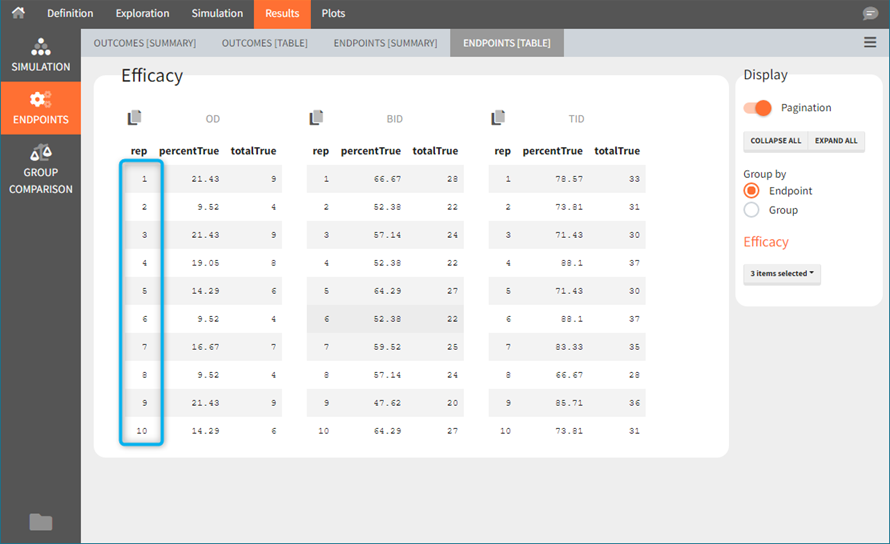
In “summaries” the values are summarized over all individuals and all replicates. For instance, endpoints result summarize outcomes over all replicates with the uncertainty of the result.
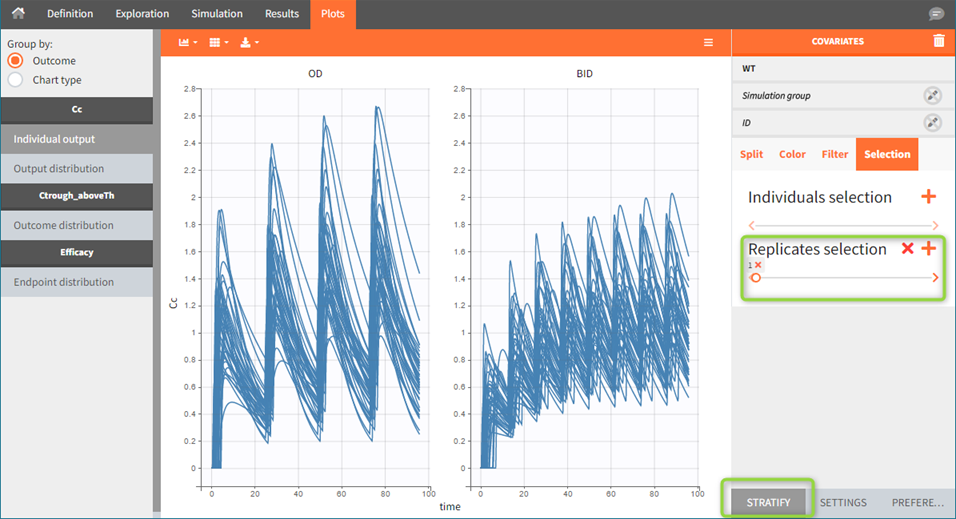
Similarly, plots contain information of all replicates. By default, individual outputs display only one replicate, however, stratification panel allows to select one or more replicate to plot (green frame).
Endpoint distribution in case of replicates is summarized in a box plot.
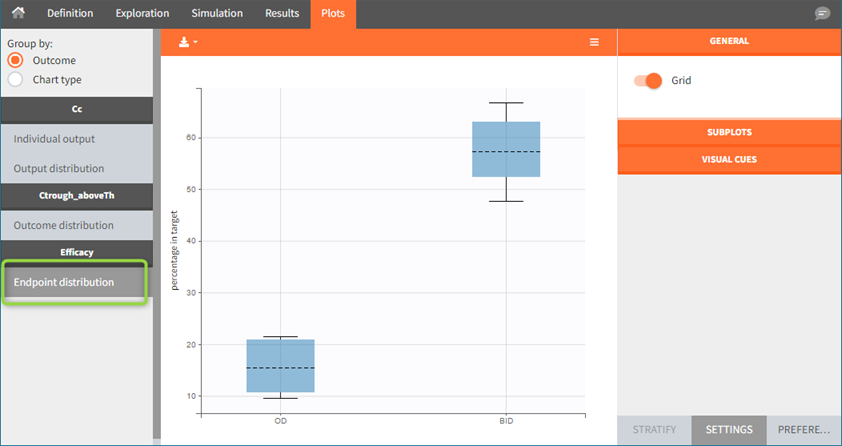
Sampling options
Individual sampling
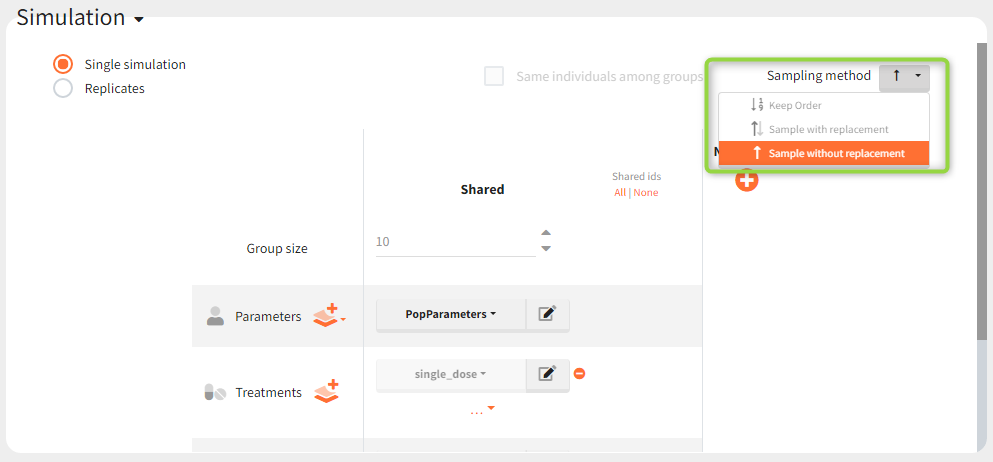
https://www.youtube.com/watch?v=q0JInVT9m3kSimulation elements of different types are subject to different individual sampling in a simulation. Elements defined as vectors provide common values for all individuals, so there is no sampling. When an element is a distribution, then sampling is done for each individual separately. Tables, from external files or from Monolix after importing a project from Monolix, can contain data for more then one individual distinguished with the column called “id”. In this case, there are three sampling methods available in the Simulation tab.
Keep order (default) – individual values are taken in the same order as they appear in a table.
With replacement – individual values are samples from a table with replacement.
Without replacement – individual values are samples from a table without replacement. This option is available only if tables contain at least the same number of individual values as a group size.
All of the above sampling methods are general and apply to all tables in a simulation scenario.
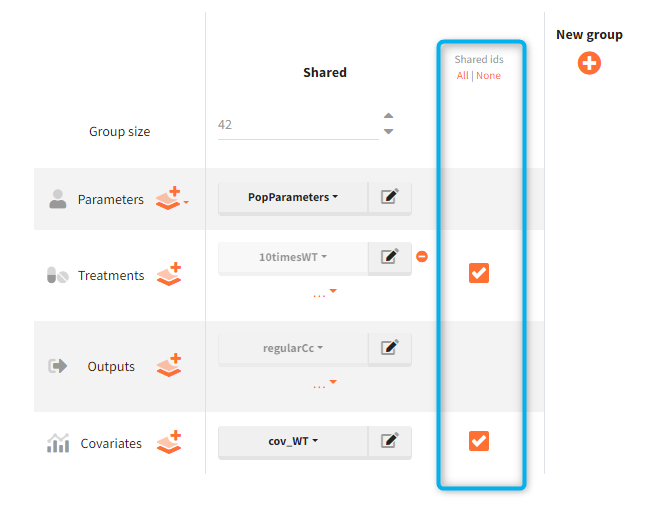
Shared ids
https://www.youtube.com/watch?v=1FbpE7tdzDoIf there is a group of tables with individual values, an option “shared ids” allows to create an intersections of ids present in the considered tables. After that, ids from these intersection are sampled to create the data for simulation.
Shared IDs checkboxes are automatically selected when individual parameter elements are combined with the covariate element “mlx_Cov” in the simulation setup, to link covariate information with the corresponding individual.
Example: Simulation uses a treatment with a dose amount depending on the patients weights. Treatment element and weights are external tables with columns (id, time, amount) for the treatment, and (id, weight) for the covariate. Moreover, an effect of the weight is added on the definition of the individual parameter (volume of the central compartment) in the model. It this case, it is important to match a sample from the treatment element with a sample from the covariate element. It is done by ticking two boxes in the “shared ids” column for both elements (blue frame).
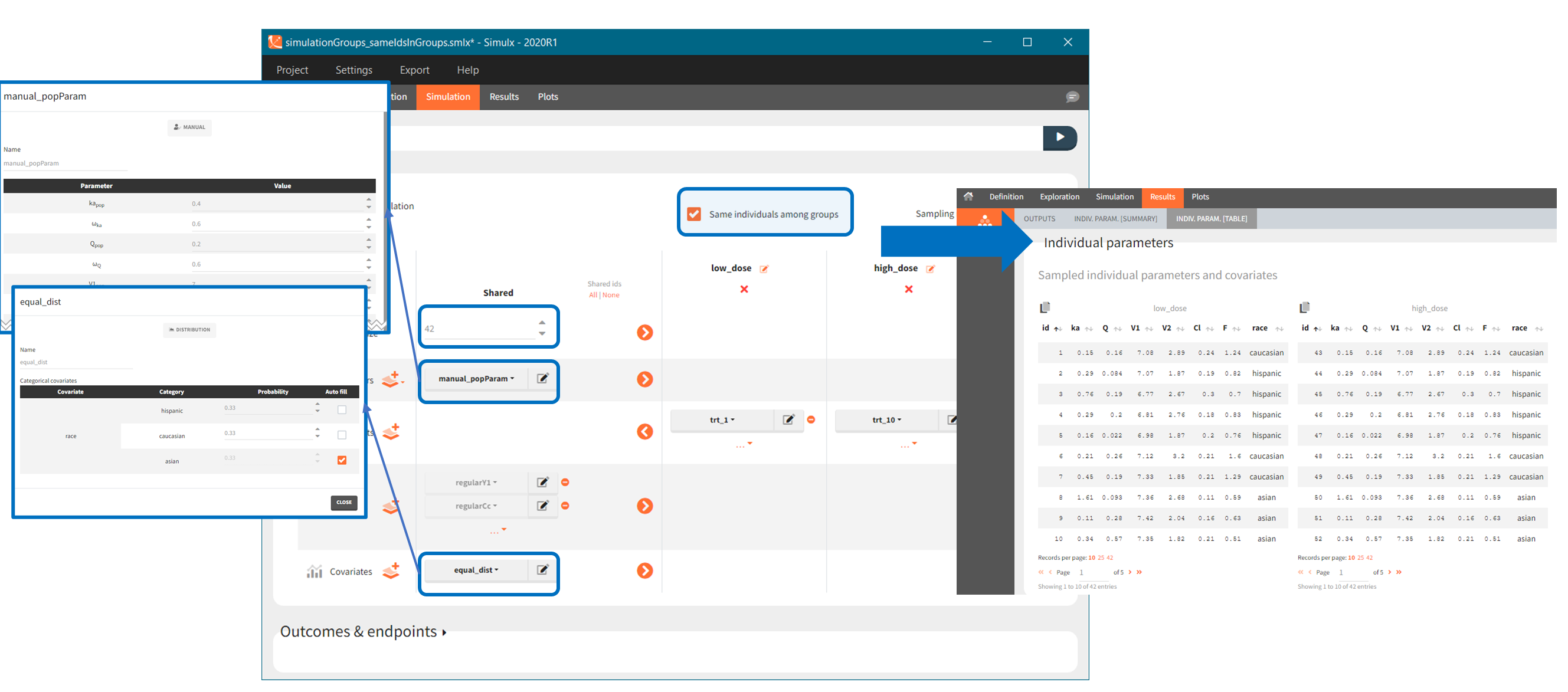
Same individuals among groups
https://www.youtube.com/watch?v=Pjmzf5kmekY“Same individuals among groups” option allows to have the same individual parameters in all groups and is available if the following elements (required for the sampling) are the same for all groups: size of groups, parameters (population or individual) and covariates. The main aim is to make the comparison between groups easier. In particular, it is used to compare different treatments on the same individuals – subjects with the same individual parameters. Selecting same individuals among groups assures that the differences between groups are only due to the treatment itself. To obtain the same conclusion without this option enabled, simulation should be performed on a very large number of individuals to averaged out the individual differences.
Example: a simulation of two treatment groups with shared parameters, covariates and outputs. When the option “same individuals among groups” is enabled, then subjects in each group have the same individual parameters because the same Parameters (manual_popParam) and Covariates (race) are used in all groups (Demo 5.simulation: simulationGroups_sameIdsInGroups).