VPCs for every event for repeated time-to-event data
In case of repeated time-to-event data, Monolix VPC plot shows the Kaplan-Meier (KM) for the first event, as well as the mean number of events per subject (cumulative plot).
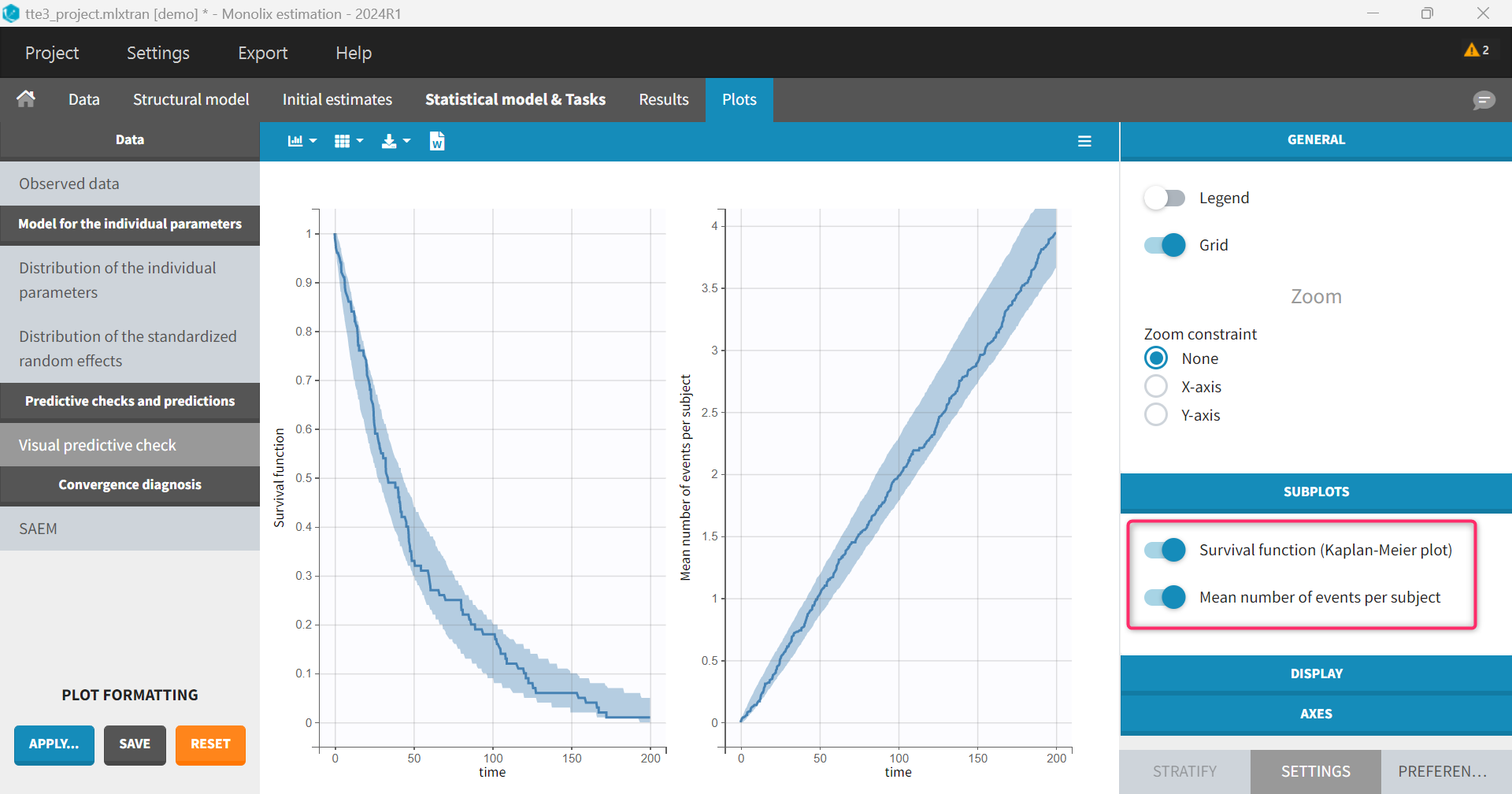
It is not possible to display in the GUI the KM plot for the second or third event for instance. However, this can be done in R.
The general workflow is the following:
use the
lixoftConnectorsto load the projectexport the VPC simulations using
computeChartsData(plot="vpc", exportVPCSimulations = T)[this step can be skipped if the VPC simulations are exported via the GUI using Export > Export VPC simulations]read the original data as well as the simulated data
filter the original and simulate data to keep only the nth event
use
survfitfrom thesurvivalpackage to generate a KM curve for the original data and each replicate of the simulated data. A time grid must be specified in order to get the KM curve on the same times for all replicates.calculate percentiles over replicates for the simulated data
plot the original KM curve and prediction interval using
ggplot2
The Surv() function needs to be called in different ways depending if we are considering exact events (using the Kaplan-Meier estimator) or interval censored events (using the Turnbull estimator). The code below recovers the type (exact or interval censored) information from the Monolix project and adapt the Surv() call accordingly.
library(lixoftConnectors)
initializeLixoftConnectors(software="monolix", force=T)
library(survival)
library(dplyr)
library(ggplot2)
#========= input settings
# Monolix project
proj <- paste0(getDemoPath(),"/3.models_for_noncontinuous_outcomes/3.3.time_to_event_data_model/tte3_project.mlxtran")
# number of events to display
nmax <- 9
#========================
# load project using lixoftConnectors
loadProject(proj)
# get observation name for the events
types <- getObservationInformation()$type
eventObsName <- names(which(types=="event"))
eventType <- getObservationInformation()$detailedType[eventObsName][[1]]
# export VPC simulations (can also be done via the GUI)
computeChartsData(plot="vpc", exportVPCSimulations = T)
# read file containing simulated data
sim_data <- read.csv(paste0(tools::file_path_sans_ext(proj),"/ChartsData/VisualPredictiveCheck/",eventObsName,"_simulations.txt"))
names(sim_data)[4] <- "events" # replacing name with eventObsName
names(sim_data)[2] <- "id" # replacing name of ID column
# read file containing original data (dataset)
obs_data <- getObservationInformation()[eventObsName][[1]]
names(obs_data)[3] <- "events"
if(eventType=="intervalCensoredEvents"){
# split time column into time1 (start of interval) and time2 (end of interval)
sim_data <- sim_data %>% group_by(rep,id) %>% mutate(time1 = lag(time), time2 = time) %>% filter(row_number()!=1) %>% ungroup()
obs_data <- obs_data %>% group_by(id) %>% mutate(time1 = lag(time), time2 = time) %>% filter(row_number()!=1) %>% ungroup()
}
# max number of events
df_nmax <- sim_data %>% group_by(rep, id) %>% summarise(nbEvents=sum(events)) %>% ungroup() %>% summarise(nbEventsMax = max(nbEvents)) %>% ungroup()
nmax_sim <- df_nmax$nbEventsMax
# containers
predInterv_allEvents <- NULL
KMoriginal_allEvents <- NULL
KMcurvesdf_allEvents <- NULL
# time grid to calculate the curves
number_of_data_points <- 100
timeGrid <- sort(unique(c(obs_data$time,
seq(0, max(obs_data$time), by=max(obs_data$time)/(number_of_data_points-1)))))
# for each nth events
for(i in 1:nmax_sim){
print(paste0("Calculating KM for the ",i, "th events out of ",nmax_sim))
if(eventType=="exactEvents"){
# for each id and rep, filter to keep first row (DV=0), and nth event (row n+1)
sim_nth <- sim_data %>% group_by(rep,id) %>% filter(row_number()==1 | row_number()==min(i+1,n())) %>% ungroup()
obs_nth <- obs_data %>% group_by(id) %>% filter(row_number()==1 | row_number()==min(i+1,n())) %>% ungroup()
# create KM curve for each replicate (can be extended to do each replicate and each stratification group)
# it is important to add the "times" argument to get all KM curves on the same time grid (in order to calculate percentiles later)
KMcurves <- summary(survfit(Surv(time, events) ~ rep, data = sim_nth), times = timeGrid, extend = TRUE)
# KM curve for original data
KMcurveOriginal <- summary(survfit(Surv(time, events) ~ 1, data = obs_nth), times = timeGrid, extend = TRUE)
}else if(eventType=="intervalCensoredEvents"){
# for each id and rep, keep interval containing ith event
sim_nth <- sim_data %>% group_by(rep,id) %>% filter(row_number()==(which(cumsum(events)>=i)[1])) %>% ungroup()
obs_nth <- obs_data %>% group_by(id) %>% filter(row_number()==(which(cumsum(events)>=i)[1])) %>% ungroup()
# create KM curve for each replicate (can be extended to do each replicate and each stratification group)
# it is important to add the "times" argument to get all KM curves on the same time grid (in order to calculate percentiles later)
# the interval in which teh event has happened in [time1, time2]. No need to have a "status" argument.
KMcurves <- summary(survfit(Surv(time=time1, time2=time2, type = "interval2") ~ rep, data = sim_nth), times = timeGrid, extend = TRUE)
# same for original data
KMcurveOriginal <- summary(survfit(Surv(time=time1, time2=time2, type = "interval2") ~ 1, data = obs_nth), times = timeGrid, extend = TRUE)
}
# survfit summary object to data.frame object
KMcurves_df <- do.call(data.frame, lapply(c("time","surv","strata") , function(x) KMcurves[x])) %>% mutate(EventNumber=paste0(i,"th event"))
# calculate percentiles for each time point
predInter <- KMcurves_df %>% group_by(time) %>% summarise(p05 = quantile(surv,0.05,type=5),
p95 = quantile(surv,0.95,type=5)) %>% ungroup() %>% mutate(EventNumber=paste0(i,"th event"))
# create KM curve for original data
KMcurveOriginal_df <- do.call(data.frame, lapply(c("time","surv") , function(x) KMcurveOriginal[x])) %>% mutate(EventNumber=paste0(i,"th event"))
# bind together
KMcurvesdf_allEvents <- rbind(KMcurvesdf_allEvents,KMcurves_df)
predInterv_allEvents <- rbind(predInterv_allEvents,predInter)
KMoriginal_allEvents <- rbind(KMoriginal_allEvents,KMcurveOriginal_df)
}
# filter to keep only the number of events to display (nmax setting from user input)
KMoriginal_allEvents <- KMoriginal_allEvents %>% filter(as.numeric(substr(EventNumber,1,nchar(EventNumber)-8))<=nmax)
predInterv_allEvents <- predInterv_allEvents %>% filter(as.numeric(substr(EventNumber,1,nchar(EventNumber)-8))<=nmax)
# EventNumber as factor
KMoriginal_allEvents$EventNumber <- as.factor(KMoriginal_allEvents$EventNumber)
predInterv_allEvents$EventNumber <- as.factor(predInterv_allEvents$EventNumber)
# plot prediction interval with overlay of actual KM curve, stratified by EventNumber
ggplot()+geom_line(data=KMoriginal_allEvents, aes(x=time,y=surv), size=0.5)+
geom_ribbon(data=predInterv_allEvents, aes(x=time, ymin=p05, ymax=p95), alpha=0.5, fill="#00a4c6") +
facet_wrap(~EventNumber) + theme_bw()+
ylab("Survival") + xlab("Time")
# calculate mean number of events (to redo plot as in the GUI) as MN = sum(1-surv_i) with the sum being over i=0 to i=nmax_sim
meanNbEvents <- KMcurvesdf_allEvents %>% group_by(strata, time) %>% summarize(MN=sum(1-surv)) %>% ungroup()
meanNbEventsOrig <- KMoriginal_allEvents %>% group_by( time) %>% summarize(MN=sum(1-surv)) %>% ungroup()
# prediction interval for mean number of events
predIntervmeanNbEvents <- meanNbEvents %>% group_by(time) %>% summarise(p05 = quantile(MN,0.05,type=5),
p95 = quantile(MN,0.95,type=5)) %>% ungroup()
# plot prediction interval with overlay of actual mean number of event curve
ggplot()+geom_line(data=meanNbEventsOrig, aes(x=time,y=MN), size=0.5)+
geom_ribbon(data=predIntervmeanNbEvents, aes(x=time, ymin=p05, ymax=p95), alpha=0.5, fill="#00a4c6") +
theme_bw()+
ylab("Mean number of events") + xlab("Time") Example with exact events
Using the tte3_project.mlxtran from the demos defined in the input settings section:
#========= input settings
proj <- paste0(getDemoPath(),"/3.models_for_noncontinuous_outcomes/3.3.time_to_event_data_model/tte3_project.mlxtran")
nmax <- 9
#======================== We obtain the following plot. The plot for the 1st event is the same as the “survival function” plot in the Monolix GUI.
We also obtain the plot of the mean number of events, which is identical to the plot in the Monolix GUI:
Example with interval-censored events
This example uses the demo project PKrtte_project.mlxtran. The survival curve using the Turnbull estimator is a step function by nature. In order to have a smooth display (as in the Monolix GUI) set the step of the timeGrid setting to the same value as the intervalLength defined in the structural model.
#========= input settings
proj <- paste0(getDemoPath(),"/3.models_for_noncontinuous_outcomes/3.3.time_to_event_data_model/tte4_project.mlxtran")
nmax <- 9 #number of events to display
#======================== BELOW
nmax_sim <- 9
timeGrid <- seq(0, 60, by=5) # Set to the same value as intervalLength
# and do not plot mean number of eventsThe resulting plot is shown below. The plot for the 1st event is the same as the “survival function” plot in the Monolix GUI.
