prctilemlx
prctilemlx: prediction interval plots
Description
Compute and plot percentiles of the empirical distribution of longitudinal data. When several groups are present, the groups can be plotted as subplots or as different colors on one plot. The input can be a R object (parameter r) or a Monolix project and a variable name (arguments project and outputVariableName).
Usage
prctilemlx(
r = NULL,
col = NULL,
project = NULL,
outputVariableName = NULL,
number = 8,
level = 80,
plot = TRUE,
color = NULL,
group = NULL,
facet = TRUE,
labels = NULL,
band = NULL
)
Arguments
| a data frame with a column id, a column time and a column with values. |
| a vector with the three column indexes for id, time and y. Default = c(1,2,3). |
| simulx project filename (with extension “.smlx”) |
| name of the output to consider. By default the first output will be consider. |
| the number of intervals (i.e. the number of percentiles minus 1). |
| the largest interval (i.e. the difference between the lowest and the highest percentile). |
| if |
| colors to be used for the plots |
| variable to be used for defining groups (by default, ‘group’ is used when it exists) |
| makes subplots for different groups if |
| vector of strings |
| is deprecated (use number and level instead) ; a list with two fields
|
Details
You must define either a dataframe r (and if needed the associated col argument) or a Simulx project and the name of the output outputVariableName. The outputVariableName corresponds to the name of the model variable, not to the output element name.
Value
a ggplot object if plot=TRUE ; otherwise, a list with fields:
proba: a vector of probabilities of length
band$number+1color: a vector of colors used for the plot of length
band$numbery: a data frame with the values of the empirical percentiles computed at each time point
Examples
This simulx-GUI demo project contains 3 simulation groups, with a low, medium or high dose. In the Simulation tab, the output element ‘regularCc’ is selected, to output the variable ‘Cc’ on a regular time grid.
With facet=T (default), each group is displayed on a separate subplot with the different shades representing different percentiles. The largest band represents the 90% prediction interval.
project.file <- "~/../lixoft/simulx/simulx2020R1/demos/5.simulation/simulationGroups_treatment.smlx"
prctilemlx(project=project.file, outputVariableName = "Cc")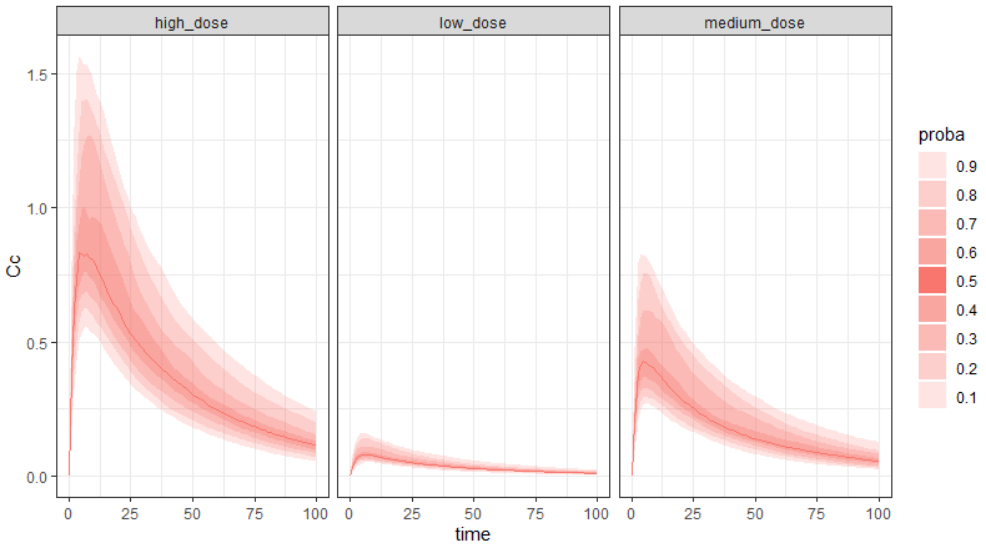
With number=2 and level=95, we can select to display only the median and the 95% prediction interval.
project.file <- "~/../lixoft/simulx/simulx2020R1/demos/5.simulation/simulationGroups_treatment.smlx"
prctilemlx(project=project.file, outputVariableName = "Cc", number=2, level=95)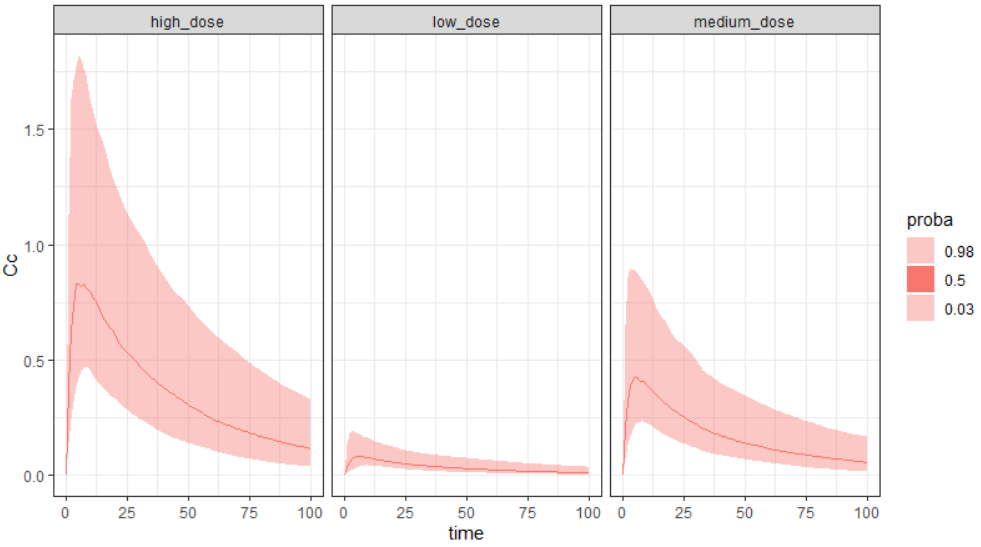
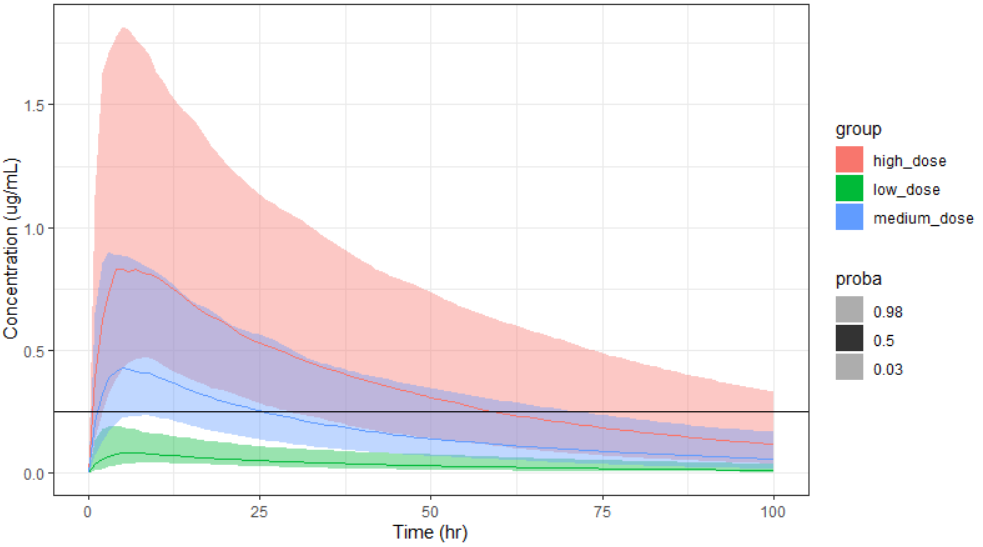
To display the prediction intervals on top of each other, use facet=F. As a ggplot object is returned, additional ggplot functions can be used to overlay additional features or modify the legend, etc.
prctilemlx(project=project.file, outputVariableName = "Cc", facet=F, number=2, level=95) + ylab("Concentration (ug/mL)") + xlab("Time (hr)") + geom_hline(yintercept = 0.25)