conc-QTc examples
Here we present in more details two examples of concentration-QTc analysis using the provided R package.
Dofetilide example
Original data
In this example, we use the data presented in Johannesen et al. and available for download. It comprises the 4 compounds dofetilide, quinidine, ranolazine, and verapamil. The clinical 5-way crossover study administered single doses of 500 µg (dofetilide), 400 mg (quinidine sulfate), 1500 mg (ranolazine), 120 mg (verapamil hydrochloride), or placebo to 22 healthy subjects. PK and QT data were collected at 16 matching time points over 24 h after dose administration.
Data processing
In order to use the provided R functions, the R script mlxQTcTools.R must be sourced. In addition, a monolixSuite installation must be available and the lixoftConnectors R package installed. In order to indicate the location of the MonolixSuite installation, the functioninitQTc(path=...) is used.
source('../_mlxQTcTools/mlxQTcTools.R')
initQTc(path="C:/Program Files/Lixoft/MonolixSuite2024R1")In this example, we will focus on the Dofetilide and Placebo data. Ths first step is thus to read the original excel dataset, filter it and save it as csv.
data <- openxlsx::read.xlsx("SCR002_clinicalData4drugs_originalFile.xlsx")
Dofetilide_data <- data %>% filter(EXTRT=="Placebo" | EXTRT=="Dofetilide") %>% select(-c(ARMCD))
write.csv(Dofetilide_data, file = "Dofetilide_data.csv", quote=F, row.names = F, na=".") The csv file contains the following columns:
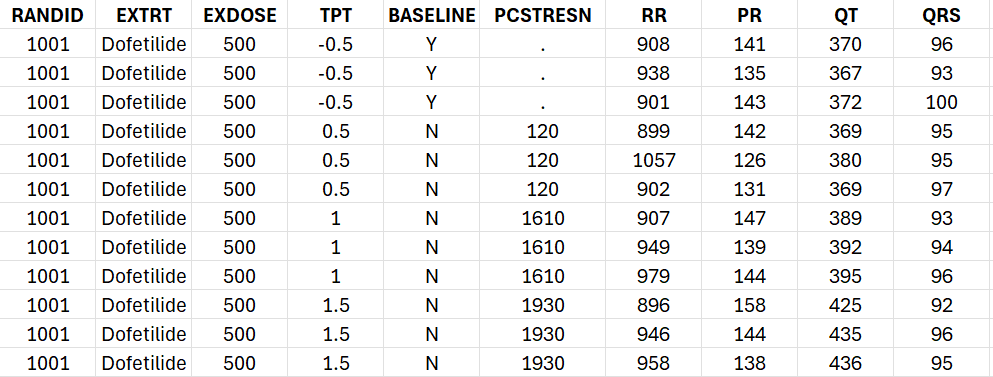
In order to perform a concentration-QTc analysis, it is necessary to apply the following processing:
calculate heart-rate correction for QT to generate QTc, for instance using Fridericia’s formula
average triplicate QTc for each time point
calculate the baseline-corrected QTc, i.e ΔQTc. In our case, the baseline is simply the pre-dose measument at time -0.5 hr. It is indicated in the dataset with BASELINE=Y
given that both placebo and dofetilide data is available for each individual, it is also possible to calculate directly the baseline-corrected and placebo-corrected QTC, i.e ΔΔQTc
for exploratory data visualization purpose, the ΔHR and ΔΔHR can also be computed
Covariates and regressor variables which will appear in the conc-QTc model are also added as additional columns:
the centered QTc baseline, and placebo-adjusted centered baseline in case of ΔΔQTc
time as categorical factor
concentration and RR interval duration
The steps can be performed using the function process_QTcData():
process_QTcData(data="Dofetilide_data.csv",
QTname = "QT", RRname = "RR", CONCname = "PCSTRESN", TRTname = "EXTRT", IDname = "RANDID", TIMEname = "TPT",
bComputeQTc = TRUE, correctionMethod = "Fridericia",
bComputeBaseline = TRUE, BLFLAGname = "BASELINE",
stepOrder = "FirstCorrectThenAverage", outName = "Dofetilide_formatted.csv", silent=F)The column containing the QT, RR, drug concentration, treatment (placebo or drug), subject id and time are indicated using the arguments QTname, RRname, CONCname, TRTname, IDname, TIMEname. Note that the values present in the TRTname column will be used as plot labels for the report. In this example, the column contains “Placebo” and “Dofetilide”. If several dose levels are used, the TRTname column should contain different values for the different dose levels, for instance “Placebo”, “Dofetilide 250 ug” and “Dofetilide 500 ug”.
If the QTc column (heart rate-corrected QT) is not present in the dataset, it can be computed using bComputeQTc = TRUE. Several correction methods are available and can be chosen using correctionMethod = "Fridericia", "Bazett", "Framingham" or "Hodges". If the QTc column is already persent, it should be inidcated using the QTCname argument.
When several replicates per time points are present, they will be averaged. It is possible to first average triplicate and then apply the heart rate correction or do the opposite using stepOrder = "FirstCorrectThenAverage" or "FirstAverageThenCorrect".
If the QTc baseline is not present yet in the dataset, it can be computed by giving bComputeBaseline = TRUE and indicating the baseline flag column in BLFLAGname. The baseline flag column should contain “0”/”1” values or “N”/”Y” values with “Y” and “1” indicating that baseline records. If several baseline records are available for each period (i.e treatment) of each individual, the baseline values will be averaged. If the baseline is already computed, the arguments BLQTCname (QTc baseline) and BLHRname (HR baseline) must be provided.
The name of the output file is given via the outName argument.
Running the function on the Dofetilide example generate the following console output:
Warning: The column 'QT' contains non-numerical values. They will be replaced by NA.
Warning: The column 'PCSTRESN' contains non-numerical values. They will be replaced by NA.
Info: The following treatment groups have been found: Placebo, Dofetilide
Info: The HR column has not been provided and has been calculated based on the 'RR' column.
Warning: The following lines had missing concentration values and do not belong to the placebo or baseline records.
The missing concentrations have been replaced by zeros.
Row number of line(s) with missing concentration: 484, 485, 486, 628, 629, 630
Info: The missing concentrations in the placebo treatment have been replaced by zeros.
Info: The missing concentrations on rows with BASELINE=Y or 1 have been replaced by zeros.
Info: The QTc column has been calculated based on the 'QT' and 'RR' columns, using Fridericia's formula: QTc=QT/(RR/1000)^(1/3)
Info: The lines corresponding to the same id, same time and same treatment (e.g triplicates) have been averaged.
Info: The BLQTc and BLHR columns have been calculated using the lines flagged as BASELINE=Y or 1.
Info: The dQTc and dHR columns have been calculated as dQTc=QTc-BLQTc and dHR=HR-BLHR.
Info: The BLQTc_cent column has been calculated as BLQTc_cent=BLQTc-mean(BLQTc).
Info: The ddQTc, ddRR and BLQTc_centAdjPl columns have be computed by subtracting the placebo data.
Info: The Cc_reg (concentration as regressor), RR_reg (RR as regressor) and TIME_cat (TIME as categorical covariate) columns have been added.
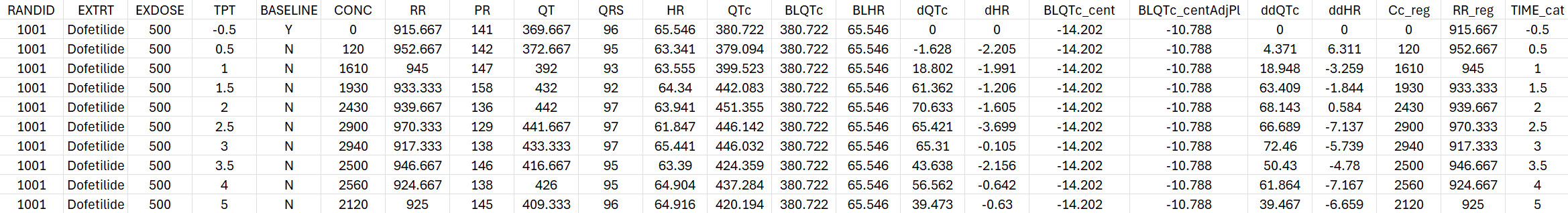
==> Created new dataset Dofetilide_formatted_.csv. The Info lines give details about the data processing steps. The Warning lines draw attention on unusual situations such as missing concentration on lines not belonging to the placebo group. The console output can be avoided using silent=T.
The generated formatted dataset is the following:
Model fitting
The next step is to generate and run Monolix projects to estimate the parameters of the concentration-QTc model(s).
Background information
Model for ΔQTc
The Garnett et al. White Paper suggests to use a linear model with concentration effect, treatment effect, time effect and centered baseline effect:
|
|
with i the individual, j the treatment and k the time. The parameters represent fixed effects and the
terms indicate random effects.
In a pharmacometric framework, this model can be rewritten as a structural model:
|
|
and a statistical model:
|
|
|
|
This formulation allows for extension to non-linear relationships between ΔQTc and the concentration, by defining alternative structural models.
Model for ΔΔQTc
ΔΔQTc can be modeled directly. With the intercept (ϴ0,pop+η0,i) and time point terms, ϴ3,T1*I(TIME=T1) + ϴ3,T2*I(TIME=T2) + ..., being identical for active and placebo treatment, these terms cancel out. The parameter ϴ1 denoting the treatment effect for ΔQTc becomes the intercept for ΔΔQTc (without random effects). A new covariate appears, the difference between centered baselines of the treatment and the placebo group. In that case, the structural model is:
|
|
The statistical model reads:
|
|
|
|
Monolix implementation
In Monolix, the structural model is defined via a txt file using mlxtran language. The concentration is passed as a regressor and its effect is defined in the structurla model. A library for model for concentration-QTc is provided with the package, including no-effect, linear, loglinear, Emax, and Emax with sigmoidicity models. Models with effect compartment to capture a delay between the concentration and the effect on QTc are also available.
The content of the mlxtran file for the linear model for ΔΔQTc is:
[LONGITUDINAL]
input = {Cc, ddQTc0, slope}
Cc = {use=regressor}
EQUATION:
ddQTc = ddQTc0 + slope*Cc
OUTPUT:
output = ddQTcThe statistical model is defined in the graphical user interface or via the lixoftConnectors.
For ΔQTc, we will consider:
an intercept
with normal distribution and random effects, and covariates TRT (categorical), TIME (categorical) and BLcent (continuous).
a slope
with normal distribution and random effects.
For ΔΔQTc, we will consider:
an intercept
without random effects and covariate BLcentAdjPl
a slope
with normal distribution and random effects.
Monolix project generation
To generate and run the Monolix projects to run the con-QTc analysis, the function generateAllQTcProjects() is used:
generateAllQTcProjects(dataSet="Dofetilide_formatted.csv",
IDname="RANDID", TIMEname="TPT", TRTname="EXTRT", CONCname="CONC", placeboName = "Placebo",
modelFolder="../_mlxQTcTools/models/ddQTcModels",
projectFolder="./mlx_runs",
isddQTC = T,
bRun = T)The user should provide the formatted dataset in the argument dataSet, as well as the name of the columns containing the subject id in IDname, time in TIMEname, treatment groups in TRTname, concentration in CONCname. The other columns (e.g QT, QTc, HR, etc) must have predefined headers and do not need to be specified.
The category of the treatment group column representing placebo can be indicated using placeboName = "Placebo". If this argument is ommited, the function will serach for the string “placebo”.
The folder containing the structural models to be used to generate the monolix runs are given in the argument modelFolder.
In folder in which the runs are generated is the projectFolder argument.
Whether conc-ΔΔQTc or conc-ΔQTc should be run is indicated using isddQTC = T or F. To run the projects, use bRun = T.
When running the function, the console output indicates the generated runs and the progress of the parameter estimation:
Created project ./mlx_runs/ddQTc_Constant.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_Emax.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_Emax_effectComp.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_EmaxSigma.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_EmaxSigma_effectComp.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_Linear.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_Linear_effectComp.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_LogLinear.mlxtran. Running project... Done.
Created project ./mlx_runs/ddQTc_LogLinear_effectComp.mlxtran. Running project... Done.The generated Monolix runs can be opened with the Monolix GUI for inspection.
Report generation
A standard report can be generated using the function quarto_render() and a Quarto template. The quarto template will read the results from generated Monolix projects and generate a report including:
Exploratory data analysis (data summary, QT/QTc, heart rate correction, baseline QTc, ΔQTc, ΔΔQTc and concentration-time)
Model assumption assessments (heart-rate independence from drug concentration, QTc independence from heart rate, linearity of the concentration-QTc relationship, immediate effect of concentration change on ΔΔQTc change)
Modeling results (comparison of the different models, model fit, parameter estimates, goodness of fit)
Derivation of the ΔΔQTc prediction intervals, including assessment of the 10-ms threshold
The quarto function is called in the following way:
quarto::quarto_render(input = "../_mlxQTcTools/QTc_quarto_allModels.qmd",
output_file = "Dofetilide_dQTc_report.docx",
execute_params = list(folderName = "../Example_Dofetilide/mlx_runs", # give relative path from quarto template (.qmd) to folder containing runs
compoundName = "Dofetilide",
CONCname = "CONC",
placeboName = "Placebo",
studyType = "dQTc",
concentrationUnit = "pg/mL",
nBins = 10,
orderTRT = c("Placebo","Dofetilide"),
thresBICc = 0))The input is the quarto template. A standard quarto template is provided in the package but it can also be adjusted to the users need. The output_file indicates the name of the generated report as word document. The execute_params list the folder containing the Monolix runs in folderName, the string to be used as compound name in compoundName, the column of the formatted dataset containing the concentration in CONCname, the placebo name in the treatment group column as argument placeboName, the type of study using studyType = "dQTc"or “ddQTc”, the concentration unit in concentrationUnit, the number of bins to average the concentration in nBins, the order of the categories present in the treatment group column to be used in plots and tables in orderTRT and the BICc threshold to consider that an alternative model is better than the linear model in thresBICc.
Download the generated ΔQTc dofetilide report and the ΔΔQTc dofetilide report.
Vanorexine example
[under construction]
