Reporting examples
NCA summary table split by a covariate horizontally
example: demo project_Theo_extravasc_SS.pkx
<lixoftPLH>
data:
task: nca
metrics: [Nobs, mean, CV, min, median, max]
parameters: [Cmax, AUClast, CLss_F, Vz_F, HL_Lambda_z]
display:
units: true
inlineUnits: true
metricsDirection: horizontal
significantDigits: 4
fitToContent: true
stratification:
state: {split: [FORM]}
splitDirection: [h]
</lixoftPLH>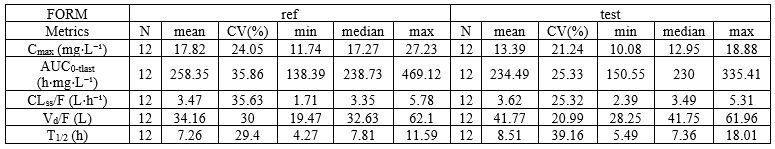
NCA summary table split by a covariate vertically
example: demo project_aPSCK9_SAD.pkx
In this example, the parameters are listed vertically, the summary metrics horizontally and the table is split by dose group vertically. The renamings are used to replace the summary metrics names.
<lixoftPLH>
data:
task: nca
metrics: [Nobs, mean, CV, min, median, max]
parameters: [AUCINF_D_obs, AUClast_D, Cl_F_obs, Cmax_D, HL_Lambda_z, Tmax, Vz_F_obs]
display:
units: true
inlineUnits: true
metricsDirection: horizontal
significantDigits: 2
fitToContent: true
stratification:
state: {split: [DOSE_mg]}
splitDirection: [v]
renamings:
mean: MEAN
Dose_mg: "Dose group"
Metrics: "Parameter"
min: MIN
median: MEDIAN
max: MAX
</lixoftPLH>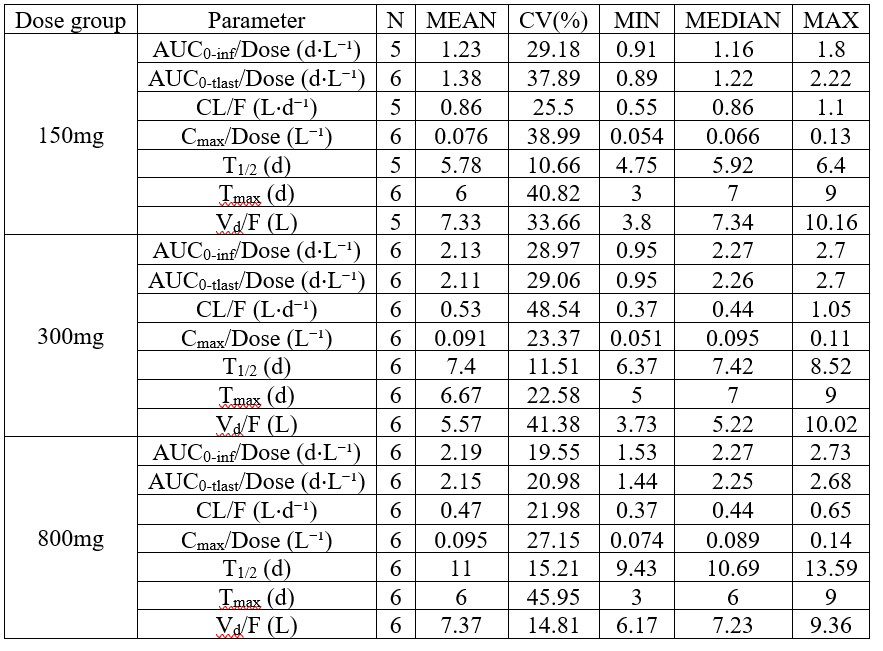
Individual NCA parameters and summary matrics in a single table
example: demo project_aPSCK9_SAD.pkx
By default when copying the tables from the GUI, individual NCA parameters and summary metrics are in two different tables but it is possible to merge them together by listing both ID and the summary metrics in the metrics argument. In this example, the table is also split by dose group and renamings are used.
<lixoftPLH>
data:
task: nca
metrics: [ID, mean, CV]
parameters: [AUCINF_D_obs, Cl_F_obs, Cmax_D, HL_Lambda_z, Tmax]
display:
units: true
inlineUnits: false
metricsDirection: vertical
significantDigits: 2
fitToContent: true
stratification:
state: {split: [DOSE_mg]}
splitDirection: [v]
renamings:
mean: MEAN
Dose_mg: "Dose group"
</lixoftPLH>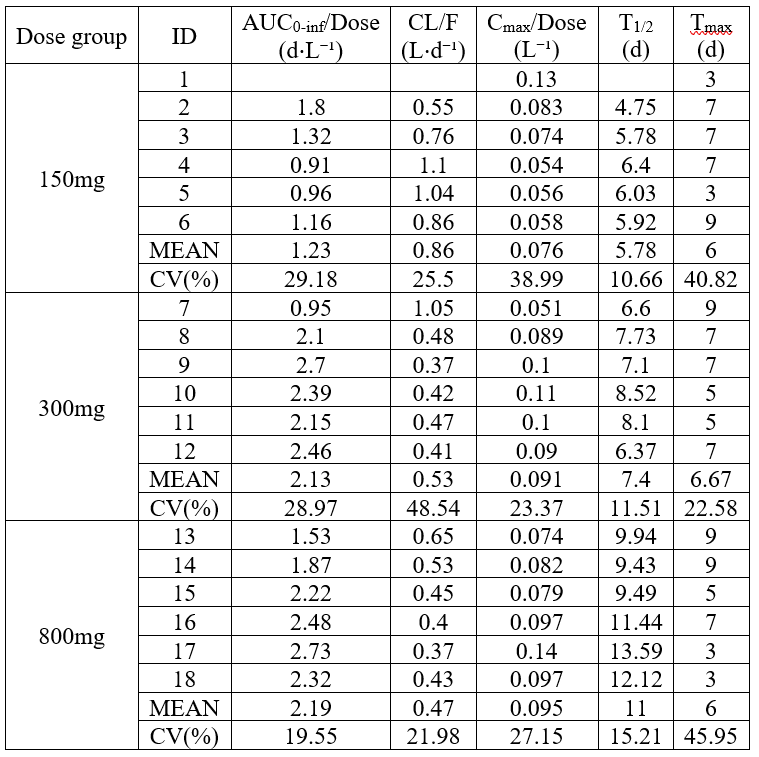
BE table for absolute bioavailability
example: demo project_parallel_absBioavailability.pkx
The default column name can be renamed using the renamings section to state “relative bioavailability” instead of “bioequivalence”.
<lixoftPLH>
data:
task: be
table: confidenceIntervals
metrics: [Ratio, CILower, CIUpper]
parameters: [AUCINF_obs, AUClast]
display:
units: true
inlineUnits: true
significantDigits: 4
fitToContent: true
renamings:
bioequivalence: "Relative bioavailability (%) based on:"
CILower: "Lower 90% CI"
CIUpper: "Upper 90% CI"
</lixoftPLH>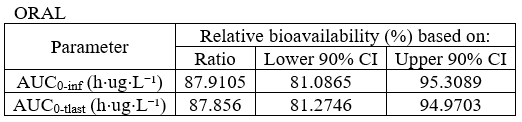
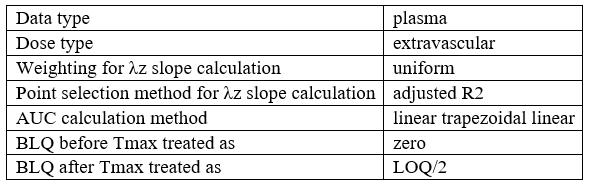
Table of NCA settings
example: demo project_censoring.pkx
Data type | %data_type% |
Dose type | %NCA_administrationType% |
Weighting for λz slope calculation | %NCA_lambdaWeighting% |
Point selection method for λz slope calculation | %NCA_lambdaRule% |
AUC calculation method | %NCA_integralMethod% |
BLQ before Tmax treated as | %NCA_blqMethodBeforeTmax% |
BLQ after Tmax treated as | %NCA_blqMethodAfterTmax% |
Individual NCA parameters for a group
example: demo DoseAndLOQ_byCategory.pkx
<lixoftPLH>
data:
task: nca
metrics: [ID]
parameters: [AUCINF_D_obs, AUCINF_obs, AUClast, AUClast_D, Cmax, Cmax_D, Tmax]
display:
units: true
inlineUnits: true
metricsDirection: vertical
significantDigits: 2
fitToContent: true
stratification:
state: {split: [STUDY], filter: [[STUDY, [1]]]}
splitDirection: [v]
</lixoftPLH>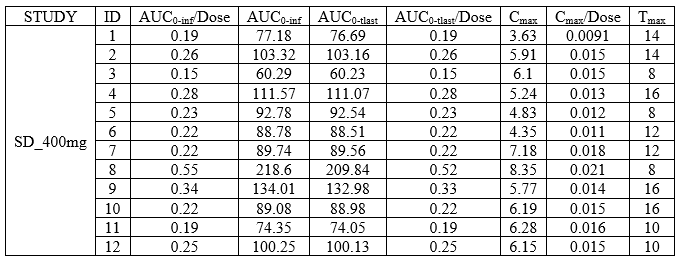
NCA summary table with summary metrics filtered by group
example: demo DoseAndLOQ_byCategory.pkx
<lixoftPLH>
data:
task: nca
metrics: [Nobs, mean, SE, CV, min, median, max, geoMean, harmMean]
parameters: [AUCINF_D_obs, AUCINF_obs, AUClast, AUClast_D, Cmax, Cmax_D, Tmax]
display:
units: true
inlineUnits: true
metricsDirection: vertical
significantDigits: 2
fitToContent: true
stratification:
state: {split: [STUDY], filter: [[STUDY, [1]]]}
splitDirection: [v]
</lixoftPLH>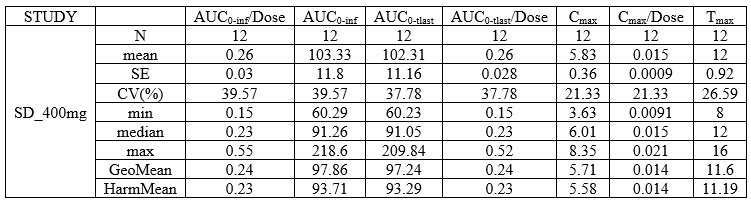
Individual covariate table
example: demo project_M2000_bolus_SD.pkx
By using excludedParameters: all, it is possible to hide all parameters and keep only covariates.
<lixoftPLH>
data:
task: nca
metrics: [ID]
excludedParameters: all
covariates: all
covariatesAfterParameters: true
display:
units: true
inlineUnits: true
metricsDirection: vertical
significantDigits: 4
fitToContent: true
</lixoftPLH>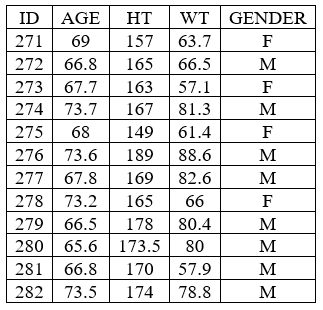
Included in LambdaZ table
example: demo project_censoring.pkx
<lixoftPLH>
data:
task: nca
table: pointsIncludedForLambdaZ
metrics: [ID, time, concentration, BLQ, includedForLambdaZ]
display:
significantDigits: 2
fitToContent: true
</lixoftPLH>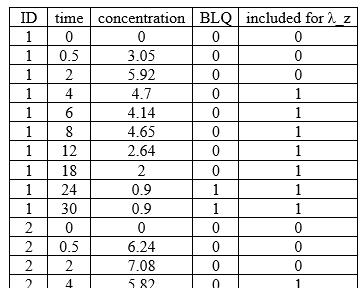
Individual NCA parameters for ref and test
example: demo project_Theo_extravasc_SD.pkx
The occasion column is shown by default but can be hidden with nbOccDisplayed: 0.
<lixoftPLH>
data:
task: nca
metrics: [ID]
parameters: [AUCINF_obs, AUClast, Cmax]
covariates: FORM
covariatesAfterParameters: false
nbOccDisplayed: 0
display:
units: true
inlineUnits: true
metricsDirection: vertical
significantDigits: 3
fitToContent: true
</lixoftPLH>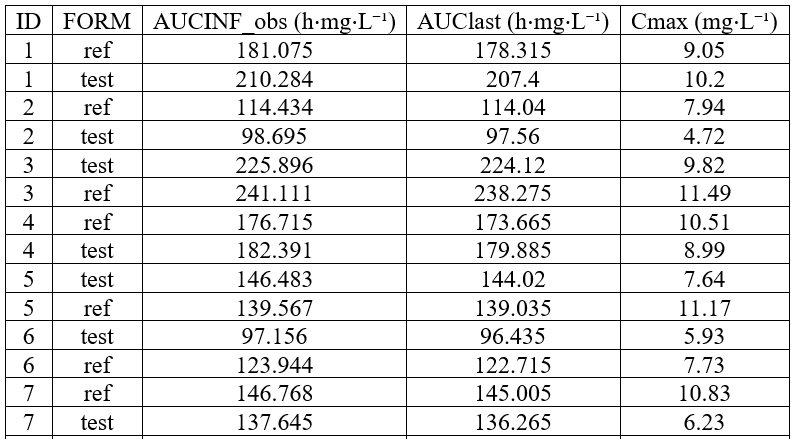
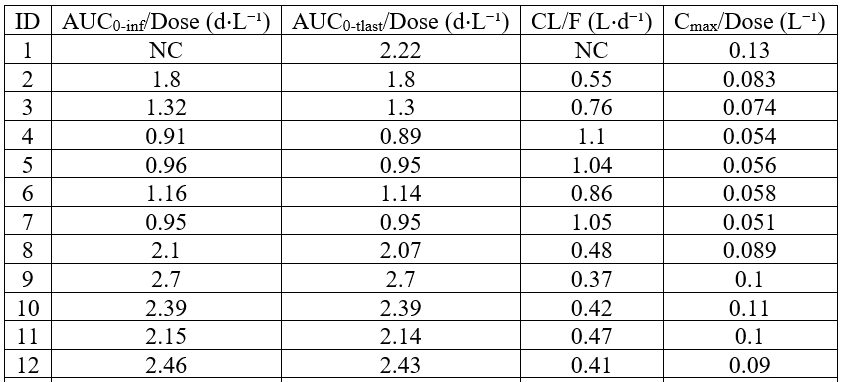
Replacing missing values by “NC”
example: demo project_aPSCK9_SAD.pkx with modified point selection for lambdaZ for id 1
By default, NCA parameters which could not be calculated are displayed as empty cell. In order to replace the empty cell by a string, it is possible to display a NaN using displayNaNs: true and then using the renamings section to replace the NaN by another string, such as NC for instance.
<lixoftPLH>
data:
task: nca
metrics: [ID]
parameters: [AUCINF_D_obs, AUClast_D, Cl_F_obs, Cmax_D]
covariates: none
covariatesAfterParameters: true
display:
units: true
inlineUnits: true
metricsDirection: vertical
significantDigits: 2
fitToContent: true
displayNaNs: true
</lixoftPLH>