Overview of the workflow
One of the main features of PKanalix is parameter estimation using a compartmental analysis framework. It estimates parameters of a compartmental model representing the PK dynamics for each individual using the Nelder-Mead algorithm. Starting with the 2023 version, PKanalix performs an individual model fit for any model with continuous outputs, either selected from the built-in library or written as a custom model.
CA task
To access the compartmental analysis task in PKanalix, in the “Tasks” tab, select the “CA” on the left-hand side, as shown below.
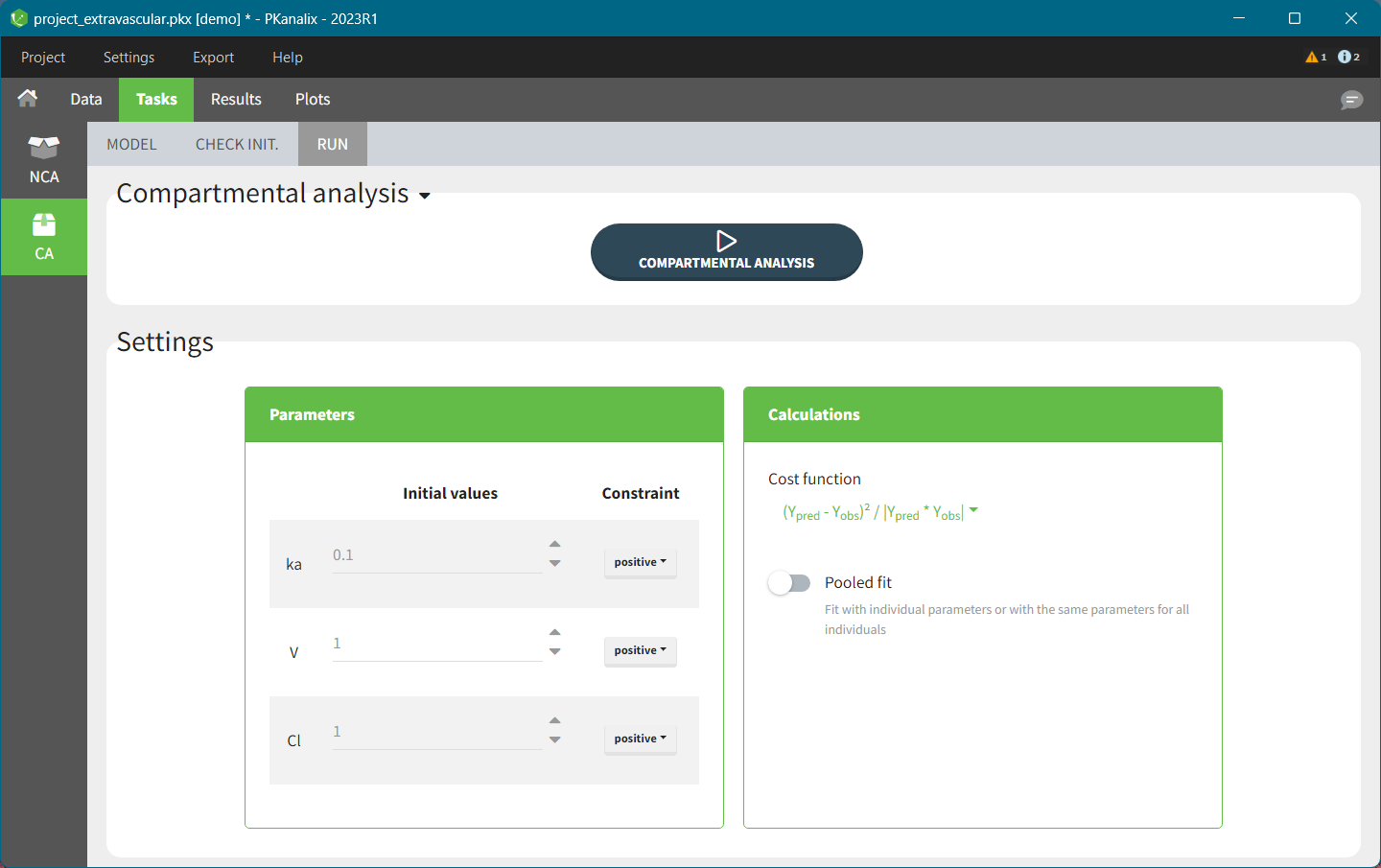
This task contains three sub-tabs, listed horizontally under the “Tasks” tab:
-
With the first, MODEL, you can select a model, e.g., from the PKanalix model library or a custom model, edit it and fit the model outputs to the observations in a dataset. For a complete guide to model definition, look at the CA model page.
-
The second, CHECK INIT., shows model predictions obtained with the initial parameter values for each individual together with the data points. It helps to select initial values for model parameters by changing values manually or using the Auto-init function, as explained in the Initialization page.
-
The third, RUN, contains a button to run the estimation algorithm and options to change the settings for the model and the estimation. The definition of all the settings and their default values is found on the Estimation page.
CA model
In this section, you specify the model. You can edit the selected model directly in the interface. In the right-hand panel,you match observations from the dataset to model outputs, see here for a complete guide.
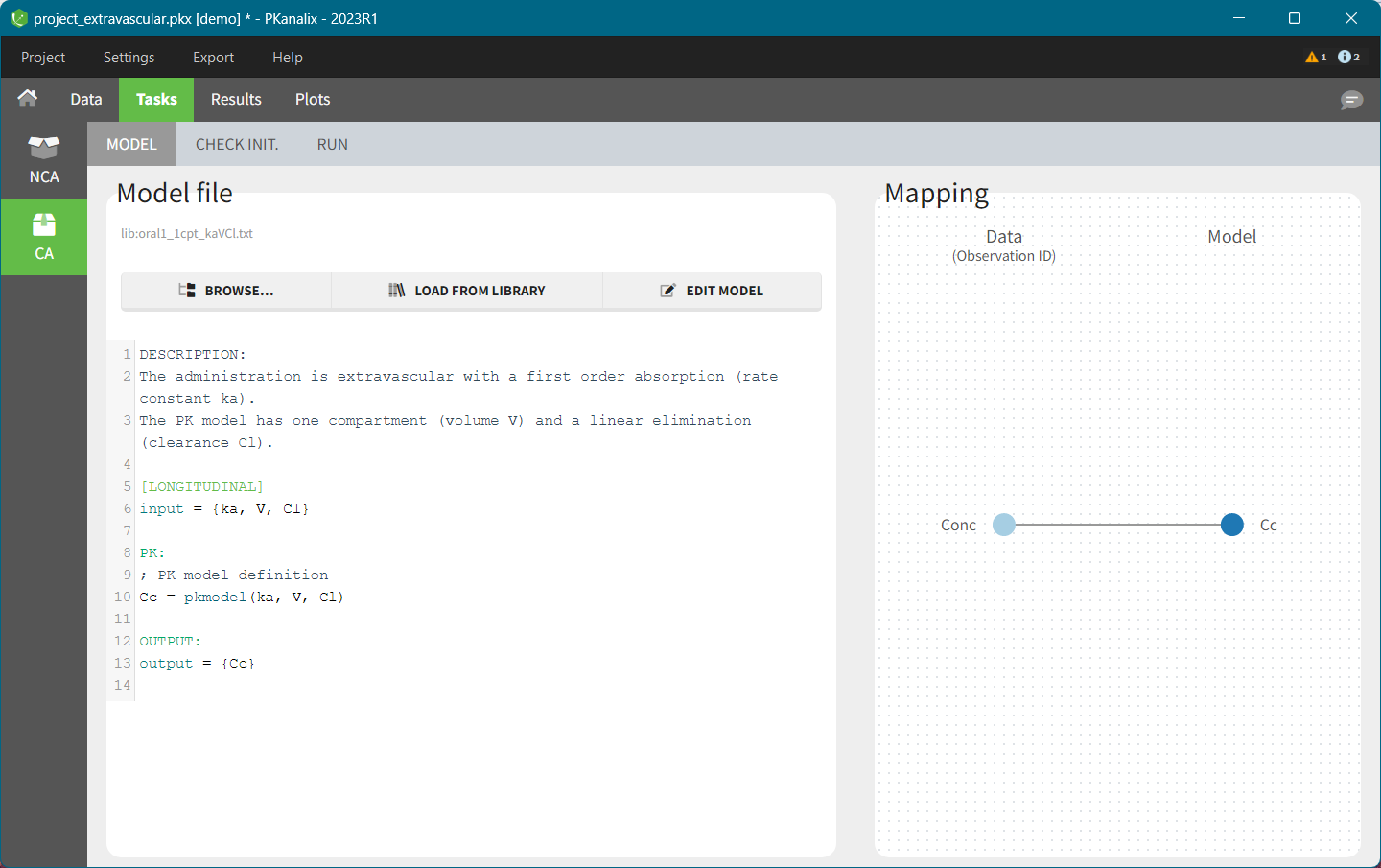
You can use custom models in PKanalix–select BROWSE and load a text file containing the model. There are also built-in libraries (PK, PK double absorption, TMDD, parent–metabolite, etc.), to simplify the modeling process. Note that models can include multiple outputs. Models used in all MonolixSuite applications are written in the mlxtran language. Note: For PKanalix versions prior to 2023, only classical PK models, from the PK model library, were available.
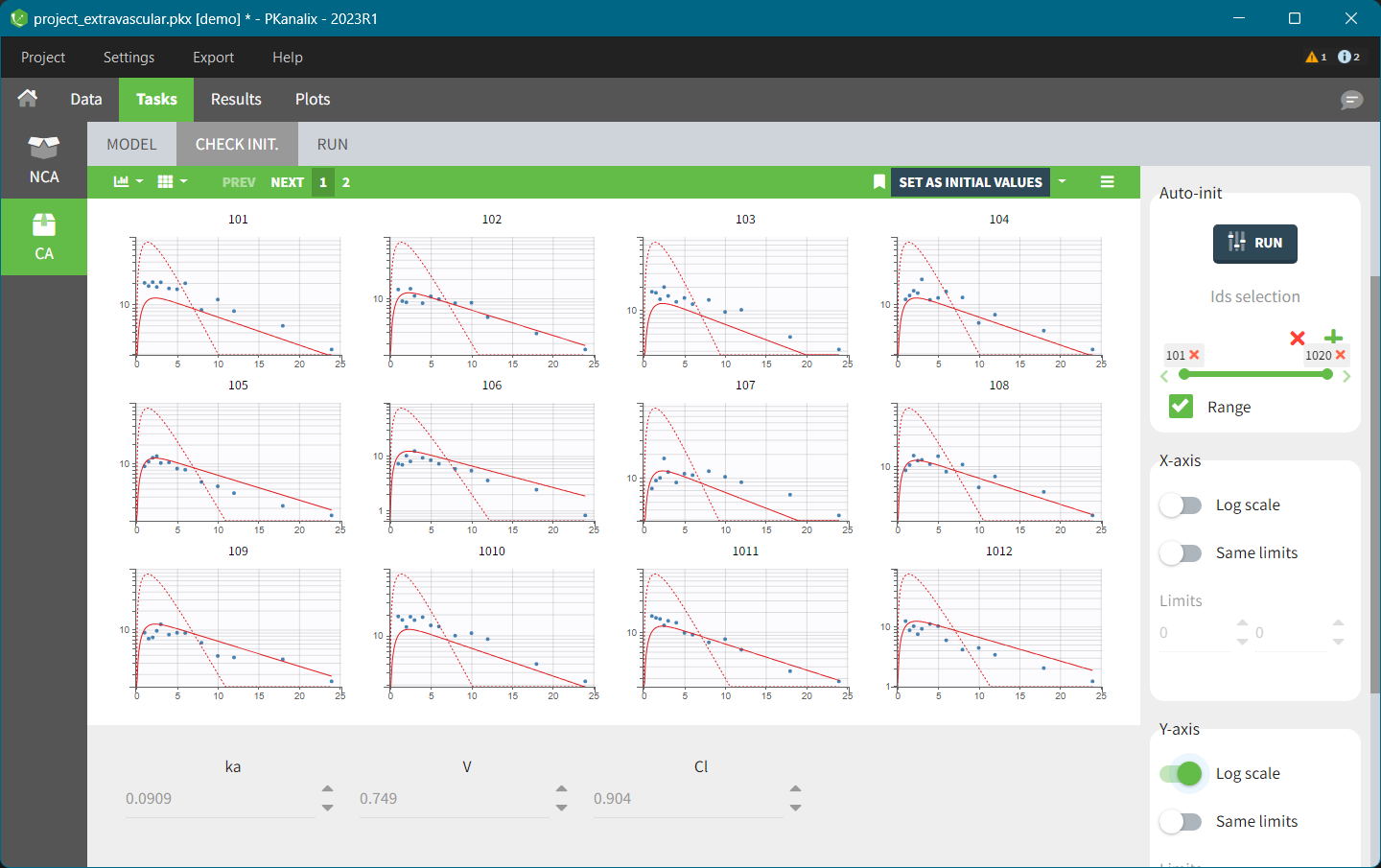
CA check init.
The “Check init.” tab helps initialize model parameters. It shows the model predictions obtained with the specified initial model parameter values and the individual designs (taking into account doses and regressors). Each sub plot shows model predictions for one individual with overlaid individual data points. You can use auto initialization to find initial values by clicking the Run button in the Auto-init panel on the right, which uses a pooled fit approach. This feature is very useful to find good initial values, as explained here.
CA results
When you run the CA task, Pkanalix automatically shows the results in the “Results” tab, see the CA Results page for a complete guide.
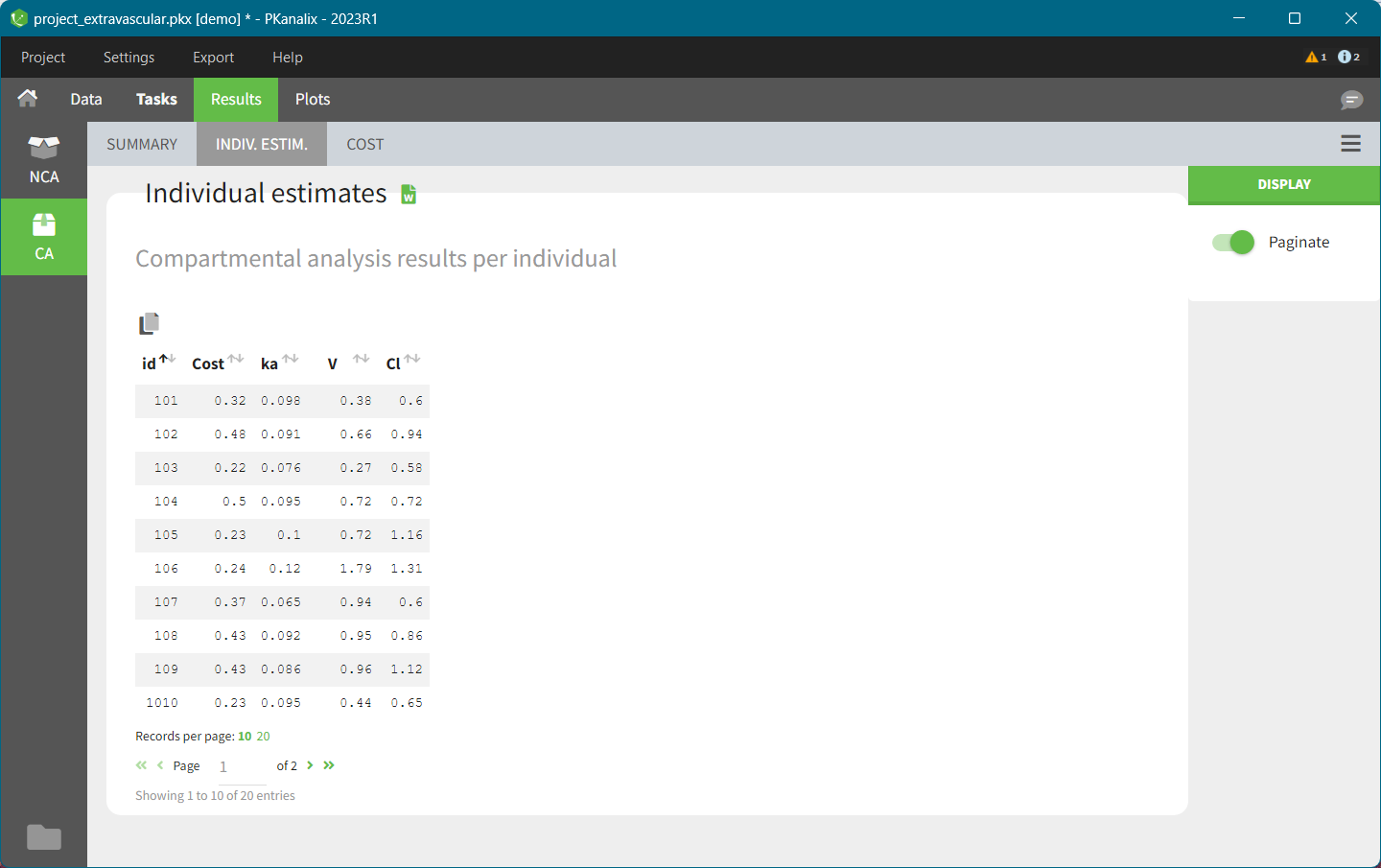
There are three main sections:
-
SUMMARY contains statistics on compartmental analysis results and summary calculation (described here). You can split or filter this table by covariates present in a dataset.
-
INDIV. ESTIM contains compartmental analysis results for each individual.
-
COST contains cost function information.
All the computed parameters depend on the chosen model. To copy a table and paste in a Word or Excel document, click on the icon on the top-left corner of a table.
CA plots
After running the CA task, several plots associated to the individual parameters are automatically generated and displayed In the “Plots” tab.
-
Individual fits: displays model fit for each individual on a separate plot.
-
Correlation between CA parameters: displays scatter plots for each pair of parameters to identify correlations between parameters.
-
Distribution of the CA parameters: displays the empirical distribution of the parameters to analyze their distribution across individuals.
-
CA parameters w.r.t. covariates: displays the individual parameters as a function of the covariates to identify correlations between the individual parameters and the covariates.
-
Observations vs predictions displays observations from a dataset versus model predictions computed from the individual parameters to detect misspecifications in the model.
CA outputs
After running the CA task, the following files are automatically generated in the project result folder/IndividualParameters/ca :
-
summary.txt contains the summary of the CA parameters calculation, in a format easily readable by a human (but not easily machine parsable).
-
cost.txt contains information about total cost and information criteria for the current model.
-
caIndividualParametersSummary.txt contains the summary of the CA parameters in a machine-friendly format useful for post-processing:
-
The first column corresponds to the name of the parameters.
-
The other columns correspond to the several elements describing the summary of the parameters (as explained here).
-
-
caIndividualParameters.txt contains the CA parameters for each subject-occasion along with the covariates.
-
The first line corresponds to the name of the parameters.
-
The other lines correspond to the value of the parameters.
-
-
You can open the files caIndividualParametersSummary.txt and caIndividualParameters.txt in R using the following command
read.table("/path/to/file.txt", sep = ",", header = TRUE)
Note: the separator used is the one defined in the user preferences. We use “,” here as it is the default.
CA export to Monolix/Simulx
Using the top menu “Export”, a PKanalix project (dataset information, model, estimated parameters) can be exported to Monolix or Simulx for further analysis, see the Export to Monolix/Simulx page for a complete guide.
-
Monolix for population modeling and further model development and diagnosis.
-
Simulx to use a CA model and estimated individual parameters for simulations of new scenarios, e.g., new dosing regimens.
Note: For PKanalix versions prior to 2023, only export to Monolix is available.

